Adarsh Krishnamurthy
Artifact Removal and Image Restoration in AFM:A Structured Mask-Guided Directional Inpainting Approach
Feb 03, 2026Abstract:Atomic Force Microscopy (AFM) enables high-resolution surface imaging at the nanoscale, yet the output is often degraded by artifacts introduced by environmental noise, scanning imperfections, and tip-sample interactions. To address this challenge, a lightweight and fully automated framework for artifact detection and restoration in AFM image analysis is presented. The pipeline begins with a classification model that determines whether an AFM image contains artifacts. If necessary, a lightweight semantic segmentation network, custom-designed and trained on AFM data, is applied to generate precise artifact masks. These masks are adaptively expanded based on their structural orientation and then inpainted using a directional neighbor-based interpolation strategy to preserve 3D surface continuity. A localized Gaussian smoothing operation is then applied for seamless restoration. The system is integrated into a user-friendly GUI that supports real-time parameter adjustments and batch processing. Experimental results demonstrate the effective artifact removal while preserving nanoscale structural details, providing a robust, geometry-aware solution for high-fidelity AFM data interpretation.
FloraForge: LLM-Assisted Procedural Generation of Editable and Analysis-Ready 3D Plant Geometric Models For Agricultural Applications
Dec 11, 2025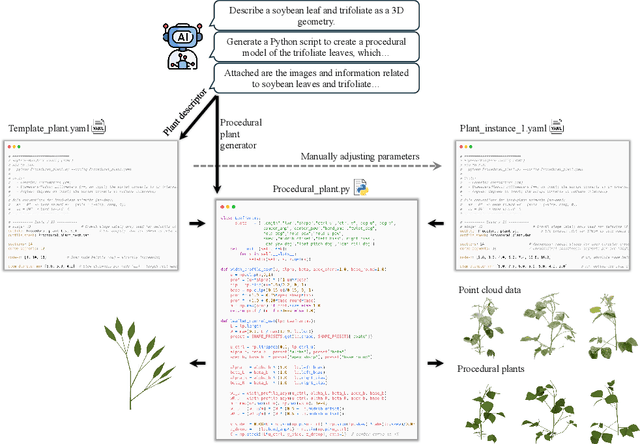
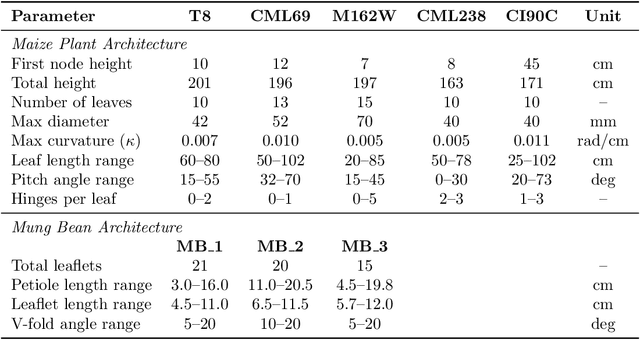
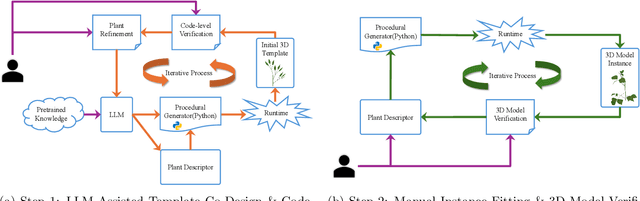
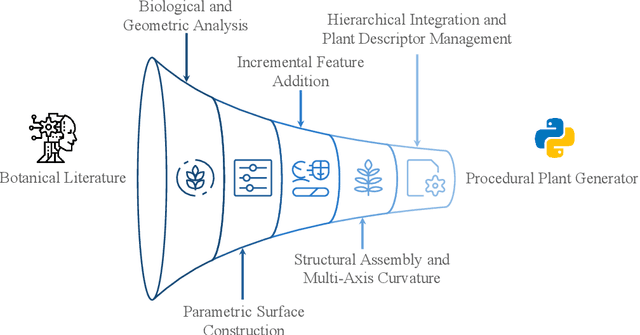
Abstract:Accurate 3D plant models are crucial for computational phenotyping and physics-based simulation; however, current approaches face significant limitations. Learning-based reconstruction methods require extensive species-specific training data and lack editability. Procedural modeling offers parametric control but demands specialized expertise in geometric modeling and an in-depth understanding of complex procedural rules, making it inaccessible to domain scientists. We present FloraForge, an LLM-assisted framework that enables domain experts to generate biologically accurate, fully parametric 3D plant models through iterative natural language Plant Refinements (PR), minimizing programming expertise. Our framework leverages LLM-enabled co-design to refine Python scripts that generate parameterized plant geometries as hierarchical B-spline surface representations with botanical constraints with explicit control points and parametric deformation functions. This representation can be easily tessellated into polygonal meshes with arbitrary precision, ensuring compatibility with functional structural plant analysis workflows such as light simulation, computational fluid dynamics, and finite element analysis. We demonstrate the framework on maize, soybean, and mung bean, fitting procedural models to empirical point cloud data through manual refinement of the Plant Descriptor (PD), human-readable files. The pipeline generates dual outputs: triangular meshes for visualization and triangular meshes with additional parametric metadata for quantitative analysis. This approach uniquely combines LLM-assisted template creation, mathematically continuous representations enabling both phenotyping and rendering, and direct parametric control through PD. The framework democratizes sophisticated geometric modeling for plant science while maintaining mathematical rigor.
Trustworthy LLM-Mediated Communication: Evaluating Information Fidelity in LLM as a Communicator (LAAC) Framework in Multiple Application Domains
Nov 06, 2025Abstract:The proliferation of AI-generated content has created an absurd communication theater where senders use LLMs to inflate simple ideas into verbose content, recipients use LLMs to compress them back into summaries, and as a consequence neither party engage with authentic content. LAAC (LLM as a Communicator) proposes a paradigm shift - positioning LLMs as intelligent communication intermediaries that capture the sender's intent through structured dialogue and facilitate genuine knowledge exchange with recipients. Rather than perpetuating cycles of AI-generated inflation and compression, LAAC enables authentic communication across diverse contexts including academic papers, proposals, professional emails, and cross-platform content generation. However, deploying LLMs as trusted communication intermediaries raises critical questions about information fidelity, consistency, and reliability. This position paper systematically evaluates the trustworthiness requirements for LAAC's deployment across multiple communication domains. We investigate three fundamental dimensions: (1) Information Capture Fidelity - accuracy of intent extraction during sender interviews across different communication types, (2) Reproducibility - consistency of structured knowledge across multiple interaction instances, and (3) Query Response Integrity - reliability of recipient-facing responses without hallucination, source conflation, or fabrication. Through controlled experiments spanning multiple LAAC use cases, we assess these trust dimensions using LAAC's multi-agent architecture. Preliminary findings reveal measurable trust gaps that must be addressed before LAAC can be reliably deployed in high-stakes communication scenarios.
ProFusion: 3D Reconstruction of Protein Complex Structures from Multi-view AFM Images
Sep 17, 2025



Abstract:AI-based in silico methods have improved protein structure prediction but often struggle with large protein complexes (PCs) involving multiple interacting proteins due to missing 3D spatial cues. Experimental techniques like Cryo-EM are accurate but costly and time-consuming. We present ProFusion, a hybrid framework that integrates a deep learning model with Atomic Force Microscopy (AFM), which provides high-resolution height maps from random orientations, naturally yielding multi-view data for 3D reconstruction. However, generating a large-scale AFM imaging data set sufficient to train deep learning models is impractical. Therefore, we developed a virtual AFM framework that simulates the imaging process and generated a dataset of ~542,000 proteins with multi-view synthetic AFM images. We train a conditional diffusion model to synthesize novel views from unposed inputs and an instance-specific Neural Radiance Field (NeRF) model to reconstruct 3D structures. Our reconstructed 3D protein structures achieve an average Chamfer Distance within the AFM imaging resolution, reflecting high structural fidelity. Our method is extensively validated on experimental AFM images of various PCs, demonstrating strong potential for accurate, cost-effective protein complex structure prediction and rapid iterative validation using AFM experiments.
Towards Large Reasoning Models for Agriculture
May 25, 2025Abstract:Agricultural decision-making involves complex, context-specific reasoning, where choices about crops, practices, and interventions depend heavily on geographic, climatic, and economic conditions. Traditional large language models (LLMs) often fall short in navigating this nuanced problem due to limited reasoning capacity. We hypothesize that recent advances in large reasoning models (LRMs) can better handle such structured, domain-specific inference. To investigate this, we introduce AgReason, the first expert-curated open-ended science benchmark with 100 questions for agricultural reasoning. Evaluations across thirteen open-source and proprietary models reveal that LRMs outperform conventional ones, though notable challenges persist, with the strongest Gemini-based baseline achieving 36% accuracy. We also present AgThoughts, a large-scale dataset of 44.6K question-answer pairs generated with human oversight and equipped with synthetically generated reasoning traces. Using AgThoughts, we develop AgThinker, a suite of small reasoning models that can be run on consumer-grade GPUs, and show that our dataset can be effective in unlocking agricultural reasoning abilities in LLMs. Our project page is here: https://baskargroup.github.io/Ag_reasoning/
NeRF-based Point Cloud Reconstruction using a Stationary Camera for Agricultural Applications
Mar 27, 2025Abstract:This paper presents a NeRF-based framework for point cloud (PCD) reconstruction, specifically designed for indoor high-throughput plant phenotyping facilities. Traditional NeRF-based reconstruction methods require cameras to move around stationary objects, but this approach is impractical for high-throughput environments where objects are rapidly imaged while moving on conveyors or rotating pedestals. To address this limitation, we develop a variant of NeRF-based PCD reconstruction that uses a single stationary camera to capture images as the object rotates on a pedestal. Our workflow comprises COLMAP-based pose estimation, a straightforward pose transformation to simulate camera movement, and subsequent standard NeRF training. A defined Region of Interest (ROI) excludes irrelevant scene data, enabling the generation of high-resolution point clouds (10M points). Experimental results demonstrate excellent reconstruction fidelity, with precision-recall analyses yielding an F-score close to 100.00 across all evaluated plant objects. Although pose estimation remains computationally intensive with a stationary camera setup, overall training and reconstruction times are competitive, validating the method's feasibility for practical high-throughput indoor phenotyping applications. Our findings indicate that high-quality NeRF-based 3D reconstructions are achievable using a stationary camera, eliminating the need for complex camera motion or costly imaging equipment. This approach is especially beneficial when employing expensive and delicate instruments, such as hyperspectral cameras, for 3D plant phenotyping. Future work will focus on optimizing pose estimation techniques and further streamlining the methodology to facilitate seamless integration into automated, high-throughput 3D phenotyping pipelines.
3D Neural Operator-Based Flow Surrogates around 3D geometries: Signed Distance Functions and Derivative Constraints
Mar 21, 2025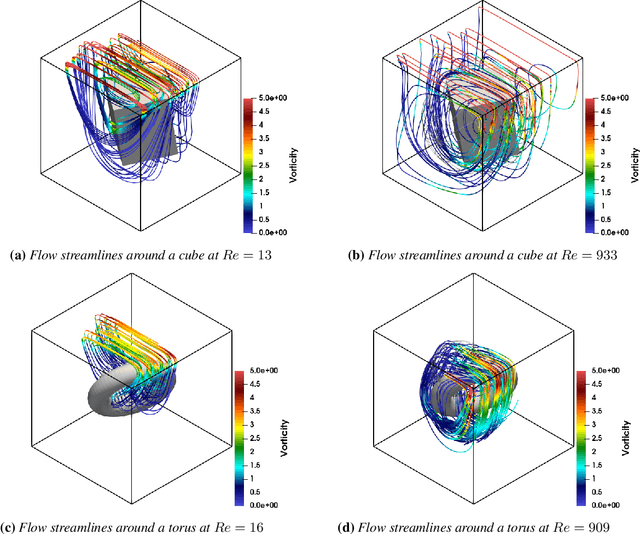
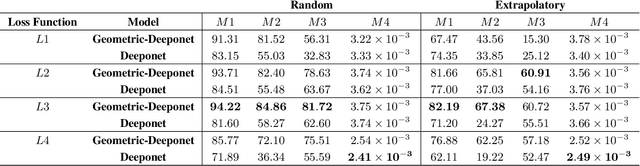
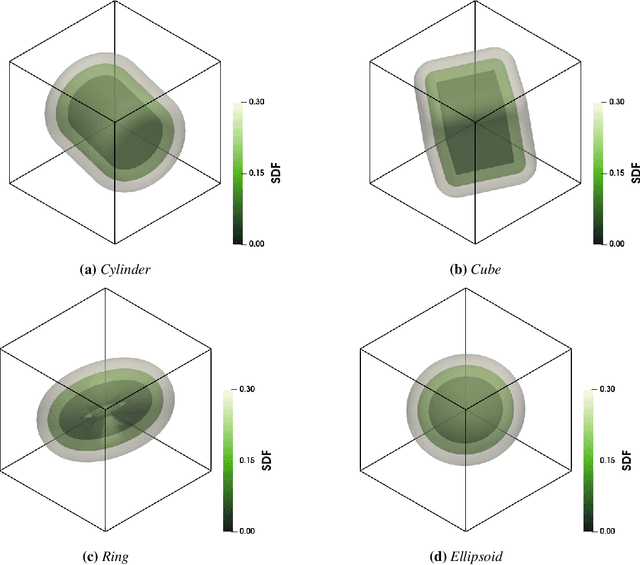
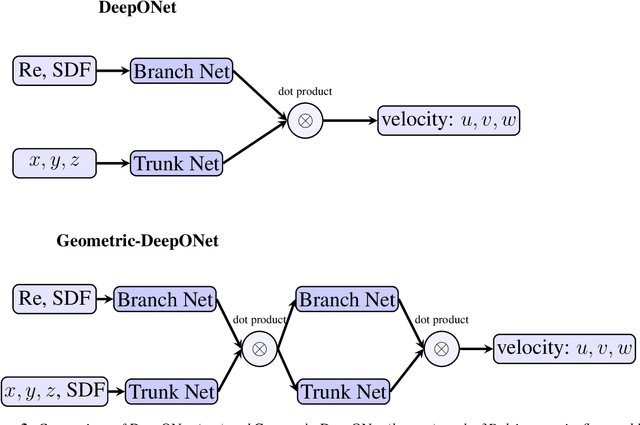
Abstract:Accurate modeling of fluid dynamics around complex geometries is critical for applications such as aerodynamic optimization and biomedical device design. While advancements in numerical methods and high-performance computing have improved simulation capabilities, the computational cost of high-fidelity 3D flow simulations remains a significant challenge. Scientific machine learning (SciML) offers an efficient alternative, enabling rapid and reliable flow predictions. In this study, we evaluate Deep Operator Networks (DeepONet) and Geometric-DeepONet, a variant that incorporates geometry information via signed distance functions (SDFs), on steady-state 3D flow over complex objects. Our dataset consists of 1,000 high-fidelity simulations spanning Reynolds numbers from 10 to 1,000, enabling comprehensive training and evaluation across a range of flow regimes. To assess model generalization, we test our models on a random and extrapolatory train-test splitting. Additionally, we explore a derivative-informed training strategy that augments standard loss functions with velocity gradient penalties and incompressibility constraints, improving physics consistency in 3D flow prediction. Our results show that Geometric-DeepONet improves boundary-layer accuracy by up to 32% compared to standard DeepONet. Moreover, incorporating derivative constraints enhances gradient accuracy by 25% in interpolation tasks and up to 45% in extrapolatory test scenarios, suggesting significant improvement in generalization capabilities to unseen 3D Reynolds numbers.
AgriField3D: A Curated 3D Point Cloud and Procedural Model Dataset of Field-Grown Maize from a Diversity Panel
Mar 10, 2025



Abstract:The application of artificial intelligence (AI) in three-dimensional (3D) agricultural research, particularly for maize, has been limited by the scarcity of large-scale, diverse datasets. While 2D image datasets are abundant, they fail to capture essential structural details such as leaf architecture, plant volume, and spatial arrangements that 3D data provide. To address this limitation, we present AgriField3D (https://baskargroup.github.io/AgriField3D/), a curated dataset of 3D point clouds of field-grown maize plants from a diverse genetic panel, designed to be AI-ready for advancing agricultural research. Our dataset comprises over 1,000 high-quality point clouds collected using a Terrestrial Laser Scanner, complemented by procedural models that provide structured, parametric representations of maize plants. These procedural models, generated using Non-Uniform Rational B-Splines (NURBS) and optimized via a two-step process combining Particle Swarm Optimization (PSO) and differentiable programming, enable precise, scalable reconstructions of leaf surfaces and plant architectures. To enhance usability, we performed graph-based segmentation to isolate individual leaves and stalks, ensuring consistent labeling across all samples. We also conducted rigorous manual quality control on all datasets, correcting errors in segmentation, ensuring accurate leaf ordering, and validating metadata annotations. The dataset further includes metadata detailing plant morphology and quality, alongside multi-resolution subsampled versions (100k, 50k, 10k points) optimized for various computational needs. By integrating point cloud data of field grown plants with high-fidelity procedural models and ensuring meticulous manual validation, AgriField3D provides a comprehensive foundation for AI-driven phenotyping, plant structural analysis, and 3D applications in agricultural research.
Accessing the Effect of Phyllotaxy and Planting Density on Light Use Efficiency in Field-Grown Maize using 3D Reconstructions
Mar 10, 2025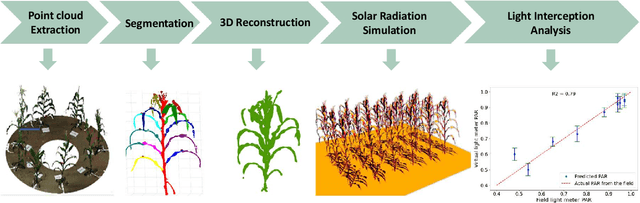
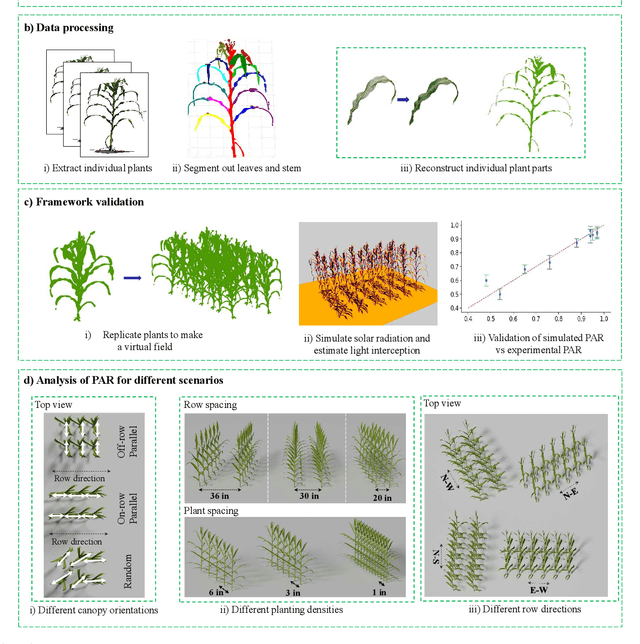
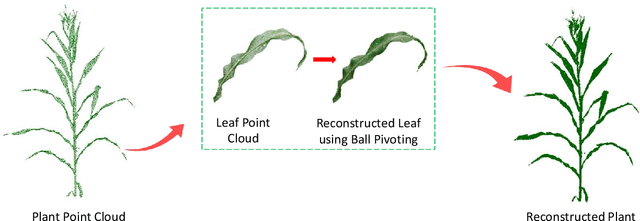
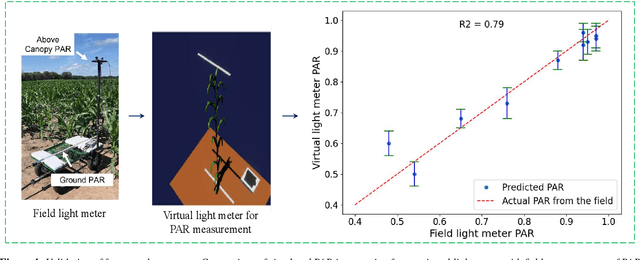
Abstract:High-density planting is a widely adopted strategy to enhance maize productivity, yet it introduces challenges such as increased interplant competition and shading, which can limit light capture and overall yield potential. In response, some maize plants naturally reorient their canopies to optimize light capture, a process known as canopy reorientation. Understanding this adaptive response and its impact on light capture is crucial for maximizing agricultural yield potential. This study introduces an end-to-end framework that integrates realistic 3D reconstructions of field-grown maize with photosynthetically active radiation (PAR) modeling to assess the effects of phyllotaxy and planting density on light interception. In particular, using 3D point clouds derived from field data, virtual fields for a diverse set of maize genotypes were constructed and validated against field PAR measurements. Using this framework, we present detailed analyses of the impact of canopy orientations, plant and row spacings, and planting row directions on PAR interception throughout a typical growing season. Our findings highlight significant variations in light interception efficiency across different planting densities and canopy orientations. By elucidating the relationship between canopy architecture and light capture, this study offers valuable guidance for optimizing maize breeding and cultivation strategies across diverse agricultural settings.
Geometry Matters: Benchmarking Scientific ML Approaches for Flow Prediction around Complex Geometries
Dec 31, 2024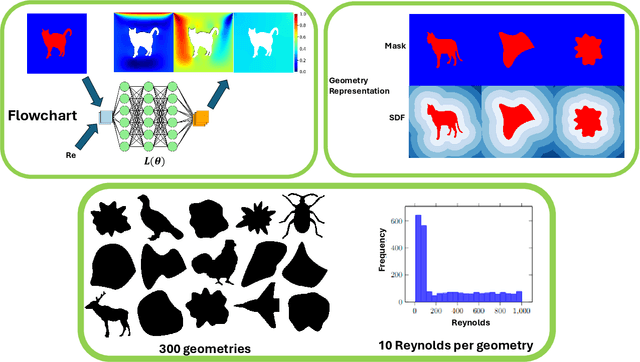
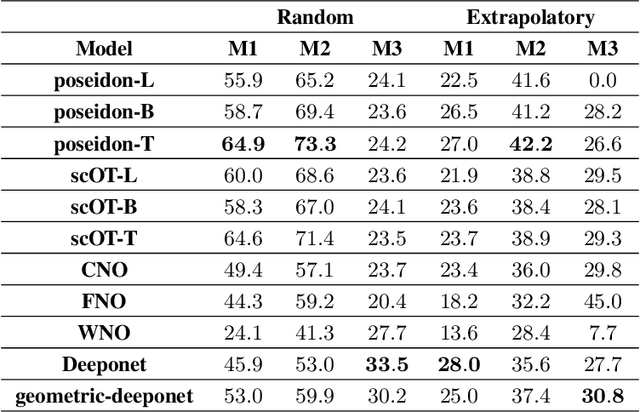
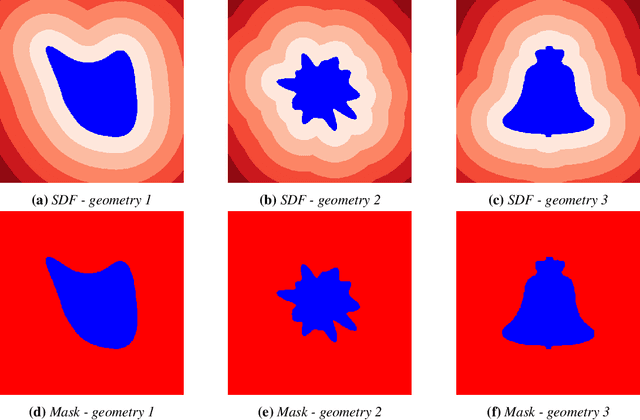
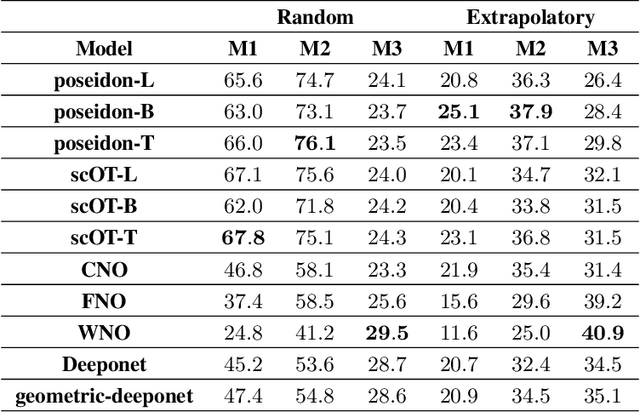
Abstract:Rapid yet accurate simulations of fluid dynamics around complex geometries is critical in a variety of engineering and scientific applications, including aerodynamics and biomedical flows. However, while scientific machine learning (SciML) has shown promise, most studies are constrained to simple geometries, leaving complex, real-world scenarios underexplored. This study addresses this gap by benchmarking diverse SciML models, including neural operators and vision transformer-based foundation models, for fluid flow prediction over intricate geometries. Using a high-fidelity dataset of steady-state flows across various geometries, we evaluate the impact of geometric representations -- Signed Distance Fields (SDF) and binary masks -- on model accuracy, scalability, and generalization. Central to this effort is the introduction of a novel, unified scoring framework that integrates metrics for global accuracy, boundary layer fidelity, and physical consistency to enable a robust, comparative evaluation of model performance. Our findings demonstrate that foundation models significantly outperform neural operators, particularly in data-limited scenarios, and that SDF representations yield superior results with sufficient training data. Despite these advancements, all models struggle with out-of-distribution generalization, highlighting a critical challenge for future SciML applications. By advancing both evaluation methodologies and modeling capabilities, this work paves the way for robust and scalable ML solutions for fluid dynamics across complex geometries.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge