Anwesha Sarkar
Artifact Removal and Image Restoration in AFM:A Structured Mask-Guided Directional Inpainting Approach
Feb 03, 2026Abstract:Atomic Force Microscopy (AFM) enables high-resolution surface imaging at the nanoscale, yet the output is often degraded by artifacts introduced by environmental noise, scanning imperfections, and tip-sample interactions. To address this challenge, a lightweight and fully automated framework for artifact detection and restoration in AFM image analysis is presented. The pipeline begins with a classification model that determines whether an AFM image contains artifacts. If necessary, a lightweight semantic segmentation network, custom-designed and trained on AFM data, is applied to generate precise artifact masks. These masks are adaptively expanded based on their structural orientation and then inpainted using a directional neighbor-based interpolation strategy to preserve 3D surface continuity. A localized Gaussian smoothing operation is then applied for seamless restoration. The system is integrated into a user-friendly GUI that supports real-time parameter adjustments and batch processing. Experimental results demonstrate the effective artifact removal while preserving nanoscale structural details, providing a robust, geometry-aware solution for high-fidelity AFM data interpretation.
ProFusion: 3D Reconstruction of Protein Complex Structures from Multi-view AFM Images
Sep 17, 2025



Abstract:AI-based in silico methods have improved protein structure prediction but often struggle with large protein complexes (PCs) involving multiple interacting proteins due to missing 3D spatial cues. Experimental techniques like Cryo-EM are accurate but costly and time-consuming. We present ProFusion, a hybrid framework that integrates a deep learning model with Atomic Force Microscopy (AFM), which provides high-resolution height maps from random orientations, naturally yielding multi-view data for 3D reconstruction. However, generating a large-scale AFM imaging data set sufficient to train deep learning models is impractical. Therefore, we developed a virtual AFM framework that simulates the imaging process and generated a dataset of ~542,000 proteins with multi-view synthetic AFM images. We train a conditional diffusion model to synthesize novel views from unposed inputs and an instance-specific Neural Radiance Field (NeRF) model to reconstruct 3D structures. Our reconstructed 3D protein structures achieve an average Chamfer Distance within the AFM imaging resolution, reflecting high structural fidelity. Our method is extensively validated on experimental AFM images of various PCs, demonstrating strong potential for accurate, cost-effective protein complex structure prediction and rapid iterative validation using AFM experiments.
3D Reconstruction of Protein Structures from Multi-view AFM Images using Neural Radiance Fields (NeRFs)
Aug 12, 2024



Abstract:Recent advancements in deep learning for predicting 3D protein structures have shown promise, particularly when leveraging inputs like protein sequences and Cryo-Electron microscopy (Cryo-EM) images. However, these techniques often fall short when predicting the structures of protein complexes (PCs), which involve multiple proteins. In our study, we investigate using atomic force microscopy (AFM) combined with deep learning to predict the 3D structures of PCs. AFM generates height maps that depict the PCs in various random orientations, providing a rich information for training a neural network to predict the 3D structures. We then employ the pre-trained UpFusion model (which utilizes a conditional diffusion model for synthesizing novel views) to train an instance-specific NeRF model for 3D reconstruction. The performance of UpFusion is evaluated through zero-shot predictions of 3D protein structures using AFM images. The challenge, however, lies in the time-intensive and impractical nature of collecting actual AFM images. To address this, we use a virtual AFM imaging process that transforms a `PDB' protein file into multi-view 2D virtual AFM images via volume rendering techniques. We extensively validate the UpFusion architecture using both virtual and actual multi-view AFM images. Our results include a comparison of structures predicted with varying numbers of views and different sets of views. This novel approach holds significant potential for enhancing the accuracy of protein complex structure predictions with further fine-tuning of the UpFusion network.
Machine learning and Topological data analysis identify unique features of human papillae in 3D scans
Jul 12, 2023Abstract:The tongue surface houses a range of papillae that are integral to the mechanics and chemistry of taste and textural sensation. Although gustatory function of papillae is well investigated, the uniqueness of papillae within and across individuals remains elusive. Here, we present the first machine learning framework on 3D microscopic scans of human papillae (n = 2092), uncovering the uniqueness of geometric and topological features of papillae. The finer differences in shapes of papillae are investigated computationally based on a number of features derived from discrete differential geometry and computational topology. Interpretable machine learning techniques show that persistent homology features of the papillae shape are the most effective in predicting the biological variables. Models trained on these features with small volumes of data samples predict the type of papillae with an accuracy of 85%. The papillae type classification models can map the spatial arrangement of filiform and fungiform papillae on a surface. Remarkably, the papillae are found to be distinctive across individuals and an individual can be identified with an accuracy of 48% among the 15 participants from a single papillae. Collectively, this is the first unprecedented evidence demonstrating that tongue papillae can serve as a unique identifier inspiring new research direction for food preferences and oral diagnostics.
3D Reconstruction of Protein Complex Structures Using Synthesized Multi-View AFM Images
Nov 26, 2022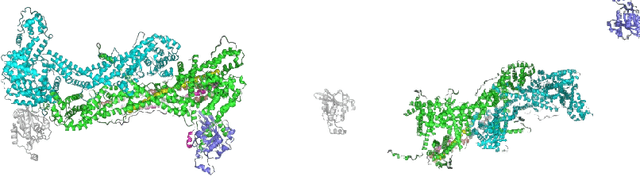


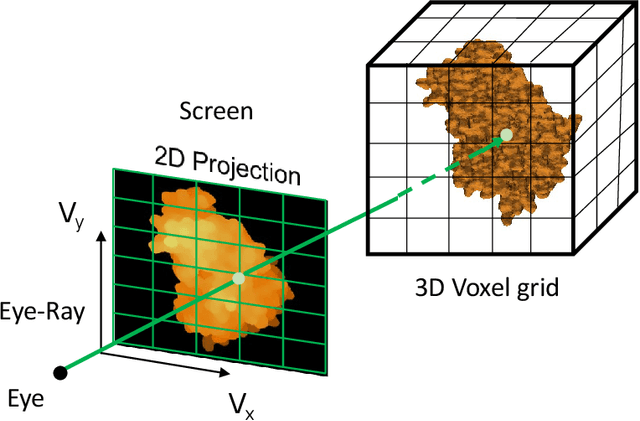
Abstract:Recent developments in deep learning-based methods demonstrated its potential to predict the 3D protein structures using inputs such as protein sequences, Cryo-Electron microscopy (Cryo-EM) images of proteins, etc. However, these methods struggle to predict the protein complexes (PC), structures with more than one protein. In this work, we explore the atomic force microscope (AFM) assisted deep learning-based methods to predict the 3D structure of PCs. The images produced by AFM capture the protein structure in different and random orientations. These multi-view images can help train the neural network to predict the 3D structure of protein complexes. However, obtaining the dataset of actual AFM images is time-consuming and not a pragmatic task. We propose a virtual AFM imaging pipeline that takes a 'PDB' protein file and generates multi-view 2D virtual AFM images using volume rendering techniques. With this, we created a dataset of around 8K proteins. We train a neural network for 3D reconstruction called Pix2Vox++ using the synthesized multi-view 2D AFM images dataset. We compare the predicted structure obtained using a different number of views and get the intersection over union (IoU) value of 0.92 on the training dataset and 0.52 on the validation dataset. We believe this approach will lead to better prediction of the structure of protein complexes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge