Soumik Sarkar
Iowa State University
ProFusion: 3D Reconstruction of Protein Complex Structures from Multi-view AFM Images
Sep 17, 2025



Abstract:AI-based in silico methods have improved protein structure prediction but often struggle with large protein complexes (PCs) involving multiple interacting proteins due to missing 3D spatial cues. Experimental techniques like Cryo-EM are accurate but costly and time-consuming. We present ProFusion, a hybrid framework that integrates a deep learning model with Atomic Force Microscopy (AFM), which provides high-resolution height maps from random orientations, naturally yielding multi-view data for 3D reconstruction. However, generating a large-scale AFM imaging data set sufficient to train deep learning models is impractical. Therefore, we developed a virtual AFM framework that simulates the imaging process and generated a dataset of ~542,000 proteins with multi-view synthetic AFM images. We train a conditional diffusion model to synthesize novel views from unposed inputs and an instance-specific Neural Radiance Field (NeRF) model to reconstruct 3D structures. Our reconstructed 3D protein structures achieve an average Chamfer Distance within the AFM imaging resolution, reflecting high structural fidelity. Our method is extensively validated on experimental AFM images of various PCs, demonstrating strong potential for accurate, cost-effective protein complex structure prediction and rapid iterative validation using AFM experiments.
Reinforcement Learning for Autonomous Point-to-Point UAV Navigation
Sep 17, 2025Abstract:Unmanned Aerial Vehicles (UAVs) are increasingly used in automated inspection, delivery, and navigation tasks that require reliable autonomy. This project develops a reinforcement learning (RL) approach to enable a single UAV to autonomously navigate between predefined points without manual intervention. The drone learns navigation policies through trial-and-error interaction, using a custom reward function that encourages goal-reaching efficiency while penalizing collisions and unsafe behavior. The control system integrates ROS with a Gym-compatible training environment, enabling flexible deployment and testing. After training, the learned policy is deployed on a real UAV platform and evaluated under practical conditions. Results show that the UAV can successfully perform autonomous navigation with minimal human oversight, demonstrating the viability of RL-based control for point-to-point drone operations in real-world scenarios.
Balancing Utility and Privacy: Dynamically Private SGD with Random Projection
Sep 11, 2025Abstract:Stochastic optimization is a pivotal enabler in modern machine learning, producing effective models for various tasks. However, several existing works have shown that model parameters and gradient information are susceptible to privacy leakage. Although Differentially Private SGD (DPSGD) addresses privacy concerns, its static noise mechanism impacts the error bounds for model performance. Additionally, with the exponential increase in model parameters, efficient learning of these models using stochastic optimizers has become more challenging. To address these concerns, we introduce the Dynamically Differentially Private Projected SGD (D2P2-SGD) optimizer. In D2P2-SGD, we combine two important ideas: (i) dynamic differential privacy (DDP) with automatic gradient clipping and (ii) random projection with SGD, allowing dynamic adjustment of the tradeoff between utility and privacy of the model. It exhibits provably sub-linear convergence rates across different objective functions, matching the best available rate. The theoretical analysis further suggests that DDP leads to better utility at the cost of privacy, while random projection enables more efficient model learning. Extensive experiments across diverse datasets show that D2P2-SGD remarkably enhances accuracy while maintaining privacy. Our code is available here.
Data-driven Kinematic Modeling in Soft Robots: System Identification and Uncertainty Quantification
Jul 10, 2025Abstract:Precise kinematic modeling is critical in calibration and controller design for soft robots, yet remains a challenging issue due to their highly nonlinear and complex behaviors. To tackle the issue, numerous data-driven machine learning approaches have been proposed for modeling nonlinear dynamics. However, these models suffer from prediction uncertainty that can negatively affect modeling accuracy, and uncertainty quantification for kinematic modeling in soft robots is underexplored. In this work, using limited simulation and real-world data, we first investigate multiple linear and nonlinear machine learning models commonly used for kinematic modeling of soft robots. The results reveal that nonlinear ensemble methods exhibit the most robust generalization performance. We then develop a conformal kinematic modeling framework for soft robots by utilizing split conformal prediction to quantify predictive position uncertainty, ensuring distribution-free prediction intervals with a theoretical guarantee.
Can Pretrained Vision-Language Embeddings Alone Guide Robot Navigation?
Jun 17, 2025



Abstract:Foundation models have revolutionized robotics by providing rich semantic representations without task-specific training. While many approaches integrate pretrained vision-language models (VLMs) with specialized navigation architectures, the fundamental question remains: can these pretrained embeddings alone successfully guide navigation without additional fine-tuning or specialized modules? We present a minimalist framework that decouples this question by training a behavior cloning policy directly on frozen vision-language embeddings from demonstrations collected by a privileged expert. Our approach achieves a 74% success rate in navigation to language-specified targets, compared to 100% for the state-aware expert, though requiring 3.2 times more steps on average. This performance gap reveals that pretrained embeddings effectively support basic language grounding but struggle with long-horizon planning and spatial reasoning. By providing this empirical baseline, we highlight both the capabilities and limitations of using foundation models as drop-in representations for embodied tasks, offering critical insights for robotics researchers facing practical design tradeoffs between system complexity and performance in resource-constrained scenarios. Our code is available at https://github.com/oadamharoon/text2nav
DeCAF: Decentralized Consensus-And-Factorization for Low-Rank Adaptation of Foundation Models
May 27, 2025Abstract:Low-Rank Adaptation (LoRA) has emerged as one of the most effective, computationally tractable fine-tuning approaches for training Vision-Language Models (VLMs) and Large Language Models (LLMs). LoRA accomplishes this by freezing the pre-trained model weights and injecting trainable low-rank matrices, allowing for efficient learning of these foundation models even on edge devices. However, LoRA in decentralized settings still remains under explored, particularly for the theoretical underpinnings due to the lack of smoothness guarantee and model consensus interference (defined formally below). This work improves the convergence rate of decentralized LoRA (DLoRA) to match the rate of decentralized SGD by ensuring gradient smoothness. We also introduce DeCAF, a novel algorithm integrating DLoRA with truncated singular value decomposition (TSVD)-based matrix factorization to resolve consensus interference. Theoretical analysis shows TSVD's approximation error is bounded and consensus differences between DLoRA and DeCAF vanish as rank increases, yielding DeCAF's matching convergence rate. Extensive experiments across vision/language tasks demonstrate our algorithms outperform local training and rivals federated learning under both IID and non-IID data distributions.
WeedNet: A Foundation Model-Based Global-to-Local AI Approach for Real-Time Weed Species Identification and Classification
May 25, 2025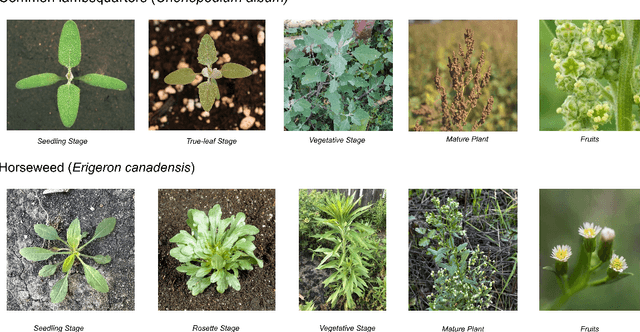
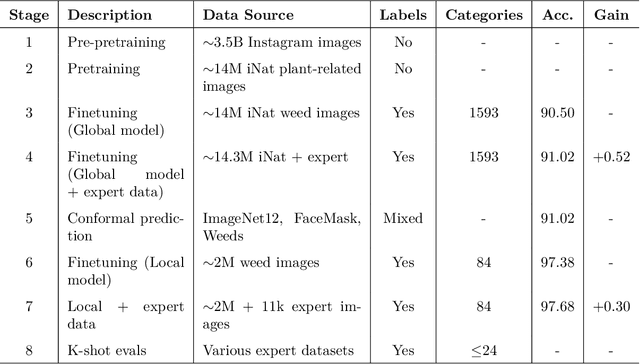
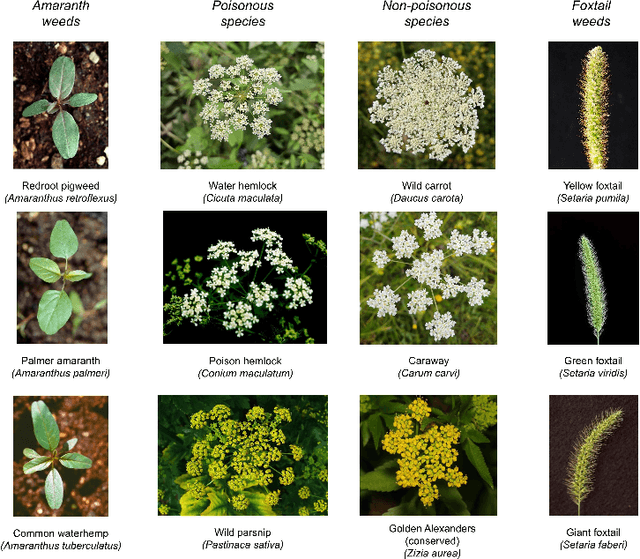

Abstract:Early identification of weeds is essential for effective management and control, and there is growing interest in automating the process using computer vision techniques coupled with AI methods. However, challenges associated with training AI-based weed identification models, such as limited expert-verified data and complexity and variability in morphological features, have hindered progress. To address these issues, we present WeedNet, the first global-scale weed identification model capable of recognizing an extensive set of weed species, including noxious and invasive plant species. WeedNet is an end-to-end real-time weed identification pipeline and uses self-supervised learning, fine-tuning, and enhanced trustworthiness strategies. WeedNet achieved 91.02% accuracy across 1,593 weed species, with 41% species achieving 100% accuracy. Using a fine-tuning strategy and a Global-to-Local approach, the local Iowa WeedNet model achieved an overall accuracy of 97.38% for 85 Iowa weeds, most classes exceeded a 90% mean accuracy per class. Testing across intra-species dissimilarity (developmental stages) and inter-species similarity (look-alike species) suggests that diversity in the images collected, spanning all the growth stages and distinguishable plant characteristics, is crucial in driving model performance. The generalizability and adaptability of the Global WeedNet model enable it to function as a foundational model, with the Global-to-Local strategy allowing fine-tuning for region-specific weed communities. Additional validation of drone- and ground-rover-based images highlights the potential of WeedNet for integration into robotic platforms. Furthermore, integration with AI for conversational use provides intelligent agricultural and ecological conservation consulting tools for farmers, agronomists, researchers, land managers, and government agencies across diverse landscapes.
Towards Large Reasoning Models for Agriculture
May 25, 2025Abstract:Agricultural decision-making involves complex, context-specific reasoning, where choices about crops, practices, and interventions depend heavily on geographic, climatic, and economic conditions. Traditional large language models (LLMs) often fall short in navigating this nuanced problem due to limited reasoning capacity. We hypothesize that recent advances in large reasoning models (LRMs) can better handle such structured, domain-specific inference. To investigate this, we introduce AgReason, the first expert-curated open-ended science benchmark with 100 questions for agricultural reasoning. Evaluations across thirteen open-source and proprietary models reveal that LRMs outperform conventional ones, though notable challenges persist, with the strongest Gemini-based baseline achieving 36% accuracy. We also present AgThoughts, a large-scale dataset of 44.6K question-answer pairs generated with human oversight and equipped with synthetically generated reasoning traces. Using AgThoughts, we develop AgThinker, a suite of small reasoning models that can be run on consumer-grade GPUs, and show that our dataset can be effective in unlocking agricultural reasoning abilities in LLMs. Our project page is here: https://baskargroup.github.io/Ag_reasoning/
Find the Fruit: Designing a Zero-Shot Sim2Real Deep RL Planner for Occlusion Aware Plant Manipulation
May 22, 2025Abstract:This paper presents an end-to-end deep reinforcement learning (RL) framework for occlusion-aware robotic manipulation in cluttered plant environments. Our approach enables a robot to interact with a deformable plant to reveal hidden objects of interest, such as fruits, using multimodal observations. We decouple the kinematic planning problem from robot control to simplify zero-shot sim2real transfer for the trained policy. Our results demonstrate that the trained policy, deployed using our framework, achieves up to 86.7% success in real-world trials across diverse initial conditions. Our findings pave the way toward autonomous, perception-driven agricultural robots that intelligently interact with complex foliage plants to "find the fruit" in challenging occluded scenarios, without the need for explicitly designed geometric and dynamic models of every plant scenario.
Zero-shot Sim-to-Real Transfer for Reinforcement Learning-based Visual Servoing of Soft Continuum Arms
Apr 23, 2025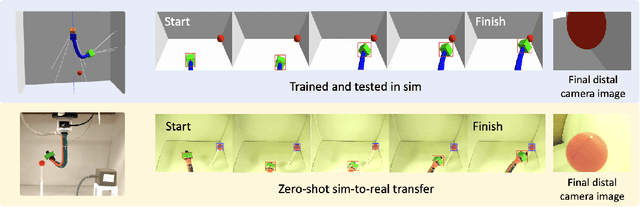
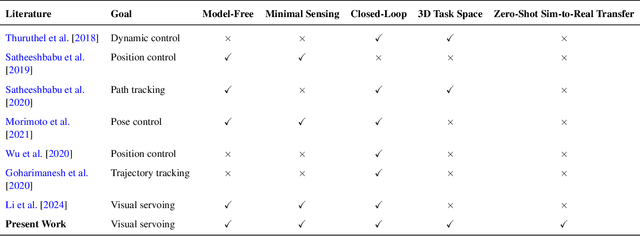
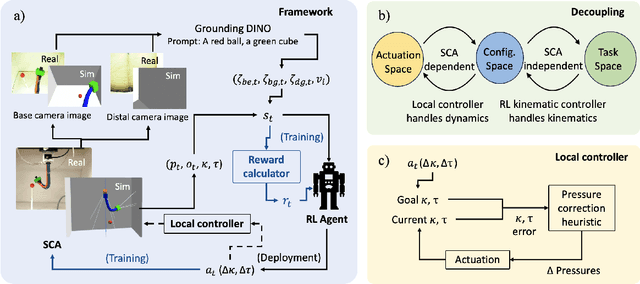
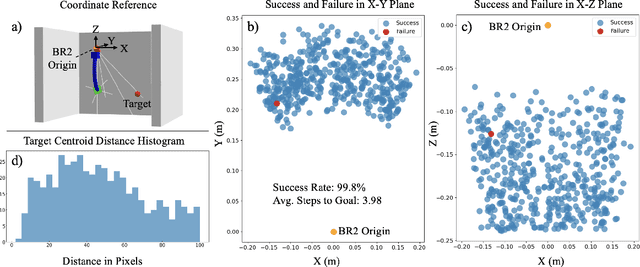
Abstract:Soft continuum arms (SCAs) soft and deformable nature presents challenges in modeling and control due to their infinite degrees of freedom and non-linear behavior. This work introduces a reinforcement learning (RL)-based framework for visual servoing tasks on SCAs with zero-shot sim-to-real transfer capabilities, demonstrated on a single section pneumatic manipulator capable of bending and twisting. The framework decouples kinematics from mechanical properties using an RL kinematic controller for motion planning and a local controller for actuation refinement, leveraging minimal sensing with visual feedback. Trained entirely in simulation, the RL controller achieved a 99.8% success rate. When deployed on hardware, it achieved a 67% success rate in zero-shot sim-to-real transfer, demonstrating robustness and adaptability. This approach offers a scalable solution for SCAs in 3D visual servoing, with potential for further refinement and expanded applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge