Talukder Jubery
AgriField3D: A Curated 3D Point Cloud and Procedural Model Dataset of Field-Grown Maize from a Diversity Panel
Mar 10, 2025



Abstract:The application of artificial intelligence (AI) in three-dimensional (3D) agricultural research, particularly for maize, has been limited by the scarcity of large-scale, diverse datasets. While 2D image datasets are abundant, they fail to capture essential structural details such as leaf architecture, plant volume, and spatial arrangements that 3D data provide. To address this limitation, we present AgriField3D (https://baskargroup.github.io/AgriField3D/), a curated dataset of 3D point clouds of field-grown maize plants from a diverse genetic panel, designed to be AI-ready for advancing agricultural research. Our dataset comprises over 1,000 high-quality point clouds collected using a Terrestrial Laser Scanner, complemented by procedural models that provide structured, parametric representations of maize plants. These procedural models, generated using Non-Uniform Rational B-Splines (NURBS) and optimized via a two-step process combining Particle Swarm Optimization (PSO) and differentiable programming, enable precise, scalable reconstructions of leaf surfaces and plant architectures. To enhance usability, we performed graph-based segmentation to isolate individual leaves and stalks, ensuring consistent labeling across all samples. We also conducted rigorous manual quality control on all datasets, correcting errors in segmentation, ensuring accurate leaf ordering, and validating metadata annotations. The dataset further includes metadata detailing plant morphology and quality, alongside multi-resolution subsampled versions (100k, 50k, 10k points) optimized for various computational needs. By integrating point cloud data of field grown plants with high-fidelity procedural models and ensuring meticulous manual validation, AgriField3D provides a comprehensive foundation for AI-driven phenotyping, plant structural analysis, and 3D applications in agricultural research.
MaizeEar-SAM: Zero-Shot Maize Ear Phenotyping
Feb 19, 2025Abstract:Quantifying the variation in yield component traits of maize (Zea mays L.), which together determine the overall productivity of this globally important crop, plays a critical role in plant genetics research, plant breeding, and the development of improved farming practices. Grain yield per acre is calculated by multiplying the number of plants per acre, ears per plant, number of kernels per ear, and the average kernel weight. The number of kernels per ear is determined by the number of kernel rows per ear multiplied by the number of kernels per row. Traditional manual methods for measuring these two traits are time-consuming, limiting large-scale data collection. Recent automation efforts using image processing and deep learning encounter challenges such as high annotation costs and uncertain generalizability. We tackle these issues by exploring Large Vision Models for zero-shot, annotation-free maize kernel segmentation. By using an open-source large vision model, the Segment Anything Model (SAM), we segment individual kernels in RGB images of maize ears and apply a graph-based algorithm to calculate the number of kernels per row. Our approach successfully identifies the number of kernels per row across a wide range of maize ears, showing the potential of zero-shot learning with foundation vision models combined with image processing techniques to improve automation and reduce subjectivity in agronomic data collection. All our code is open-sourced to make these affordable phenotyping methods accessible to everyone.
Evaluating NeRFs for 3D Plant Geometry Reconstruction in Field Conditions
Feb 15, 2024Abstract:We evaluate different Neural Radiance Fields (NeRFs) techniques for reconstructing (3D) plants in varied environments, from indoor settings to outdoor fields. Traditional techniques often struggle to capture the complex details of plants, which is crucial for botanical and agricultural understanding. We evaluate three scenarios with increasing complexity and compare the results with the point cloud obtained using LiDAR as ground truth data. In the most realistic field scenario, the NeRF models achieve a 74.65% F1 score with 30 minutes of training on the GPU, highlighting the efficiency and accuracy of NeRFs in challenging environments. These findings not only demonstrate the potential of NeRF in detailed and realistic 3D plant modeling but also suggest practical approaches for enhancing the speed and efficiency of the 3D reconstruction process.
Stochastic Conservative Contextual Linear Bandits
Mar 29, 2022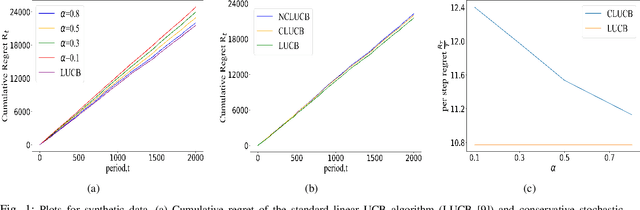
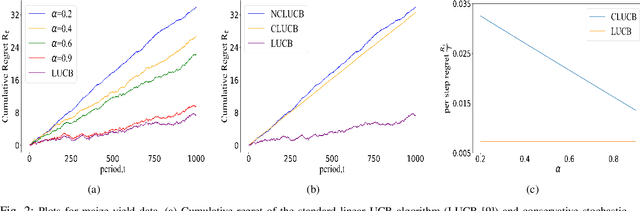
Abstract:Many physical systems have underlying safety considerations that require that the strategy deployed ensures the satisfaction of a set of constraints. Further, often we have only partial information on the state of the system. We study the problem of safe real-time decision making under uncertainty. In this paper, we formulate a conservative stochastic contextual bandit formulation for real-time decision making when an adversary chooses a distribution on the set of possible contexts and the learner is subject to certain safety/performance constraints. The learner observes only the context distribution and the exact context is unknown, and the goal is to develop an algorithm that selects a sequence of optimal actions to maximize the cumulative reward without violating the safety constraints at any time step. By leveraging the UCB algorithm for this setting, we propose a conservative linear UCB algorithm for stochastic bandits with context distribution. We prove an upper bound on the regret of the algorithm and show that it can be decomposed into three terms: (i) an upper bound for the regret of the standard linear UCB algorithm, (ii) a constant term (independent of time horizon) that accounts for the loss of being conservative in order to satisfy the safety constraint, and (ii) a constant term (independent of time horizon) that accounts for the loss for the contexts being unknown and only the distribution being known. To validate the performance of our approach we perform extensive simulations on synthetic data and on real-world maize data collected through the Genomes to Fields (G2F) initiative.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge