Talukder Zaki Jubery
Accessing the Effect of Phyllotaxy and Planting Density on Light Use Efficiency in Field-Grown Maize using 3D Reconstructions
Mar 10, 2025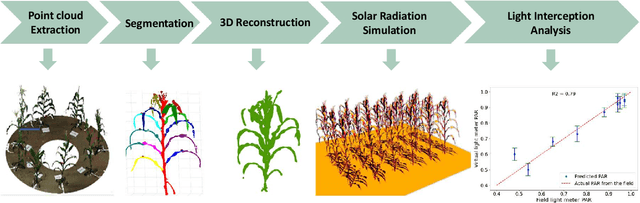
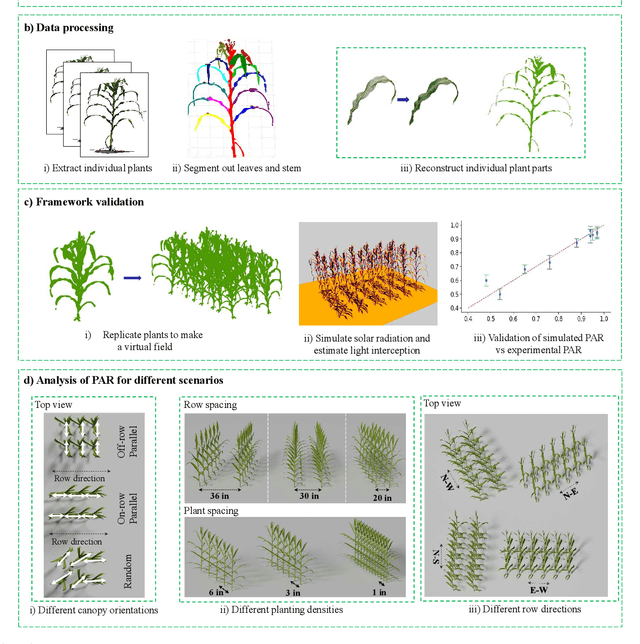
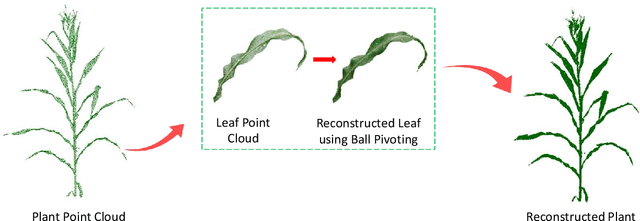
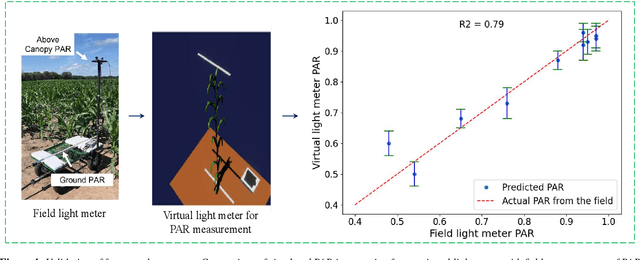
Abstract:High-density planting is a widely adopted strategy to enhance maize productivity, yet it introduces challenges such as increased interplant competition and shading, which can limit light capture and overall yield potential. In response, some maize plants naturally reorient their canopies to optimize light capture, a process known as canopy reorientation. Understanding this adaptive response and its impact on light capture is crucial for maximizing agricultural yield potential. This study introduces an end-to-end framework that integrates realistic 3D reconstructions of field-grown maize with photosynthetically active radiation (PAR) modeling to assess the effects of phyllotaxy and planting density on light interception. In particular, using 3D point clouds derived from field data, virtual fields for a diverse set of maize genotypes were constructed and validated against field PAR measurements. Using this framework, we present detailed analyses of the impact of canopy orientations, plant and row spacings, and planting row directions on PAR interception throughout a typical growing season. Our findings highlight significant variations in light interception efficiency across different planting densities and canopy orientations. By elucidating the relationship between canopy architecture and light capture, this study offers valuable guidance for optimizing maize breeding and cultivation strategies across diverse agricultural settings.
Disentangling Genotype and Environment Specific Latent Features for Improved Trait Prediction using a Compositional Autoencoder
Oct 25, 2024Abstract:This study introduces a compositional autoencoder (CAE) framework designed to disentangle the complex interplay between genotypic and environmental factors in high-dimensional phenotype data to improve trait prediction in plant breeding and genetics programs. Traditional predictive methods, which use compact representations of high-dimensional data through handcrafted features or latent features like PCA or more recently autoencoders, do not separate genotype-specific and environment-specific factors. We hypothesize that disentangling these features into genotype-specific and environment-specific components can enhance predictive models. To test this, we developed a compositional autoencoder (CAE) that decomposes high-dimensional data into distinct genotype-specific and environment-specific latent features. Our CAE framework employs a hierarchical architecture within an autoencoder to effectively separate these entangled latent features. Applied to a maize diversity panel dataset, the CAE demonstrates superior modeling of environmental influences and 5-10 times improved predictive performance for key traits like Days to Pollen and Yield, compared to the traditional methods, including standard autoencoders, PCA with regression, and Partial Least Squares Regression (PLSR). By disentangling latent features, the CAE provides powerful tool for precision breeding and genetic research. This work significantly enhances trait prediction models, advancing agricultural and biological sciences.
AgEval: A Benchmark for Zero-Shot and Few-Shot Plant Stress Phenotyping with Multimodal LLMs
Jul 29, 2024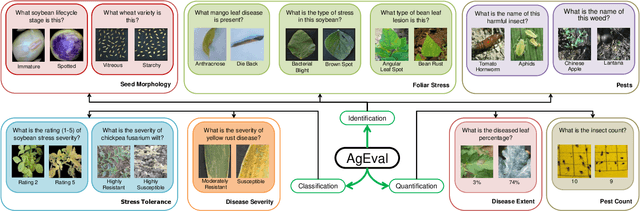
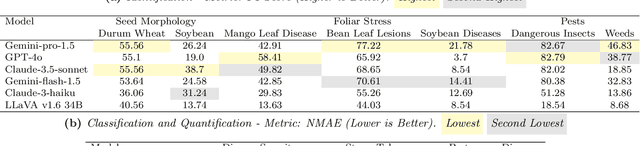
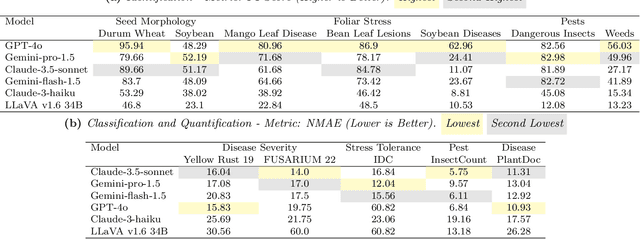
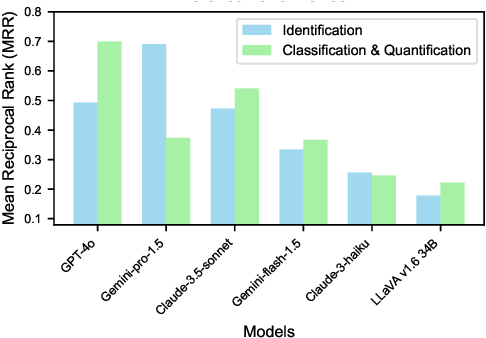
Abstract:Plant stress phenotyping traditionally relies on expert assessments and specialized models, limiting scalability in agriculture. Recent advances in multimodal large language models (LLMs) offer potential solutions to this challenge. We present AgEval, a benchmark comprising 12 diverse plant stress phenotyping tasks, to evaluate these models' capabilities. Our study assesses zero-shot and few-shot in-context learning performance of state-of-the-art models, including Claude, GPT, Gemini, and LLaVA. Results show significant performance improvements with few-shot learning, with F1 scores increasing from 46.24% to 73.37% in 8-shot identification for the best-performing model. Few-shot examples from other classes in the dataset have negligible or negative impacts, although having the exact category example helps to increase performance by 15.38%. We also quantify the consistency of model performance across different classes within each task, finding that the coefficient of variance (CV) ranges from 26.02% to 58.03% across models, implying that subject matter expertise is needed - of 'difficult' classes - to achieve reliability in performance. AgEval establishes baseline metrics for multimodal LLMs in agricultural applications, offering insights into their promise for enhancing plant stress phenotyping at scale. Benchmark and code can be accessed at: https://anonymous.4open.science/r/AgEval/
Class-specific Data Augmentation for Plant Stress Classification
Jun 18, 2024
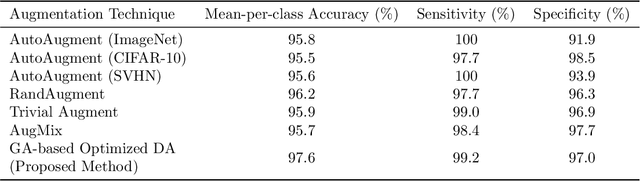
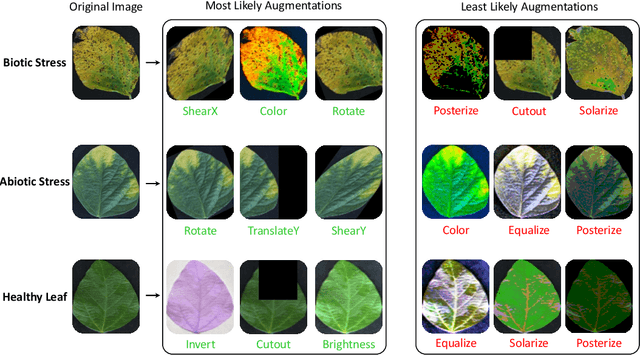
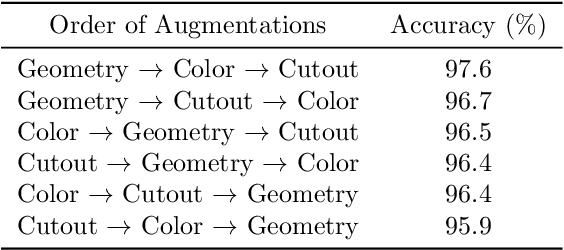
Abstract:Data augmentation is a powerful tool for improving deep learning-based image classifiers for plant stress identification and classification. However, selecting an effective set of augmentations from a large pool of candidates remains a key challenge, particularly in imbalanced and confounding datasets. We propose an approach for automated class-specific data augmentation using a genetic algorithm. We demonstrate the utility of our approach on soybean [Glycine max (L.) Merr] stress classification where symptoms are observed on leaves; a particularly challenging problem due to confounding classes in the dataset. Our approach yields substantial performance, achieving a mean-per-class accuracy of 97.61% and an overall accuracy of 98% on the soybean leaf stress dataset. Our method significantly improves the accuracy of the most challenging classes, with notable enhancements from 83.01% to 88.89% and from 85.71% to 94.05%, respectively. A key observation we make in this study is that high-performing augmentation strategies can be identified in a computationally efficient manner. We fine-tune only the linear layer of the baseline model with different augmentations, thereby reducing the computational burden associated with training classifiers from scratch for each augmentation policy while achieving exceptional performance. This research represents an advancement in automated data augmentation strategies for plant stress classification, particularly in the context of confounding datasets. Our findings contribute to the growing body of research in tailored augmentation techniques and their potential impact on disease management strategies, crop yields, and global food security. The proposed approach holds the potential to enhance the accuracy and efficiency of deep learning-based tools for managing plant stresses in agriculture.
Out-of-distribution detection algorithms for robust insect classification
May 02, 2023Abstract:Deep learning-based approaches have produced models with good insect classification accuracy; Most of these models are conducive for application in controlled environmental conditions. One of the primary emphasis of researchers is to implement identification and classification models in the real agriculture fields, which is challenging because input images that are wildly out of the distribution (e.g., images like vehicles, animals, humans, or a blurred image of an insect or insect class that is not yet trained on) can produce an incorrect insect classification. Out-of-distribution (OOD) detection algorithms provide an exciting avenue to overcome these challenge as it ensures that a model abstains from making incorrect classification prediction of non-insect and/or untrained insect class images. We generate and evaluate the performance of state-of-the-art OOD algorithms on insect detection classifiers. These algorithms represent a diversity of methods for addressing an OOD problem. Specifically, we focus on extrusive algorithms, i.e., algorithms that wrap around a well-trained classifier without the need for additional co-training. We compared three OOD detection algorithms: (i) Maximum Softmax Probability, which uses the softmax value as a confidence score, (ii) Mahalanobis distance-based algorithm, which uses a generative classification approach; and (iii) Energy-Based algorithm that maps the input data to a scalar value, called energy. We performed an extensive series of evaluations of these OOD algorithms across three performance axes: (a) \textit{Base model accuracy}: How does the accuracy of the classifier impact OOD performance? (b) How does the \textit{level of dissimilarity to the domain} impact OOD performance? and (c) \textit{Data imbalance}: How sensitive is OOD performance to the imbalance in per-class sample size?
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge