Talukder Z. Jubery
FloraForge: LLM-Assisted Procedural Generation of Editable and Analysis-Ready 3D Plant Geometric Models For Agricultural Applications
Dec 11, 2025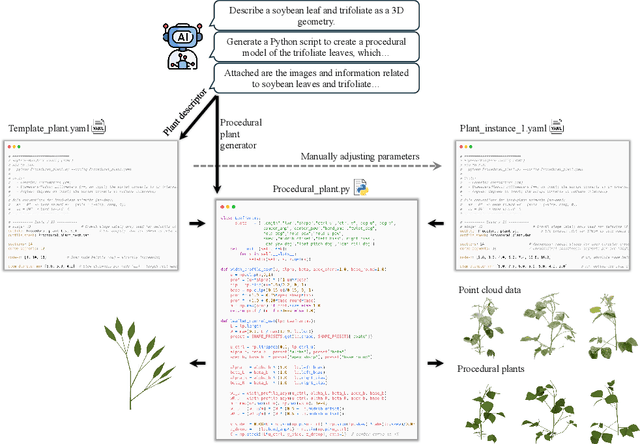
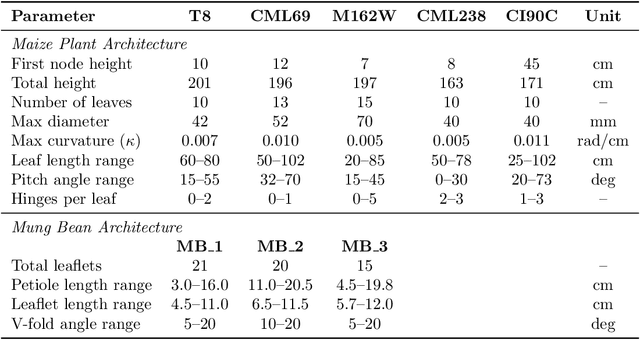
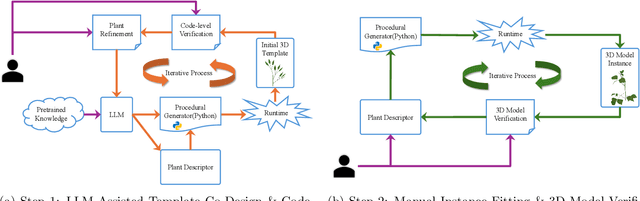
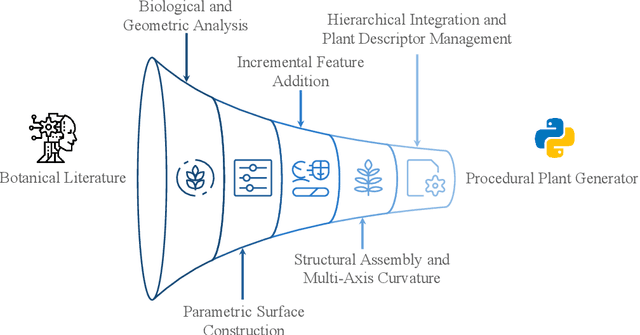
Abstract:Accurate 3D plant models are crucial for computational phenotyping and physics-based simulation; however, current approaches face significant limitations. Learning-based reconstruction methods require extensive species-specific training data and lack editability. Procedural modeling offers parametric control but demands specialized expertise in geometric modeling and an in-depth understanding of complex procedural rules, making it inaccessible to domain scientists. We present FloraForge, an LLM-assisted framework that enables domain experts to generate biologically accurate, fully parametric 3D plant models through iterative natural language Plant Refinements (PR), minimizing programming expertise. Our framework leverages LLM-enabled co-design to refine Python scripts that generate parameterized plant geometries as hierarchical B-spline surface representations with botanical constraints with explicit control points and parametric deformation functions. This representation can be easily tessellated into polygonal meshes with arbitrary precision, ensuring compatibility with functional structural plant analysis workflows such as light simulation, computational fluid dynamics, and finite element analysis. We demonstrate the framework on maize, soybean, and mung bean, fitting procedural models to empirical point cloud data through manual refinement of the Plant Descriptor (PD), human-readable files. The pipeline generates dual outputs: triangular meshes for visualization and triangular meshes with additional parametric metadata for quantitative analysis. This approach uniquely combines LLM-assisted template creation, mathematically continuous representations enabling both phenotyping and rendering, and direct parametric control through PD. The framework democratizes sophisticated geometric modeling for plant science while maintaining mathematical rigor.
Soybean Maturity Prediction using 2D Contour Plots from Drone based Time Series Imagery
Dec 12, 2024Abstract:Plant breeding programs require assessments of days to maturity for accurate selection and placement of entries in appropriate tests. In the early stages of the breeding pipeline, soybean breeding programs assign relative maturity ratings to experimental varieties that indicate their suitable maturity zones. Traditionally, the estimation of maturity value for breeding varieties has involved breeders manually inspecting fields and assessing maturity value visually. This approach relies heavily on rater judgment, making it subjective and time-consuming. This study aimed to develop a machine-learning model for evaluating soybean maturity using UAV-based time-series imagery. Images were captured at three-day intervals, beginning as the earliest varieties started maturing and continuing until the last varieties fully matured. The data collected for this experiment consisted of 22,043 plots collected across three years (2021 to 2023) and represent relative maturity groups 1.6 - 3.9. We utilized contour plot images extracted from the time-series UAV RGB imagery as input for a neural network model. This contour plot approach encoded the temporal and spatial variation within each plot into a single image. A deep learning model was trained to utilize this contour plot to predict maturity ratings. This model significantly improves accuracy and robustness, achieving up to 85% accuracy. We also evaluate the model's accuracy as we reduce the number of time points, quantifying the trade-off between temporal resolution and maturity prediction. The predictive model offers a scalable, objective, and efficient means of assessing crop maturity, enabling phenomics and ML approaches to reduce the reliance on manual inspection and subjective assessment. This approach enables the automatic prediction of relative maturity ratings in a breeding program, saving time and resources.
Multi-Sensor and Multi-temporal High-Throughput Phenotyping for Monitoring and Early Detection of Water-Limiting Stress in Soybean
Feb 28, 2024Abstract:Soybean production is susceptible to biotic and abiotic stresses, exacerbated by extreme weather events. Water limiting stress, i.e. drought, emerges as a significant risk for soybean production, underscoring the need for advancements in stress monitoring for crop breeding and production. This project combines multi-modal information to identify the most effective and efficient automated methods to investigate drought response. We investigated a set of diverse soybean accessions using multiple sensors in a time series high-throughput phenotyping manner to: (1) develop a pipeline for rapid classification of soybean drought stress symptoms, and (2) investigate methods for early detection of drought stress. We utilized high-throughput time-series phenotyping using UAVs and sensors in conjunction with machine learning (ML) analytics, which offered a swift and efficient means of phenotyping. The red-edge and green bands were most effective to classify canopy wilting stress. The Red-Edge Chlorophyll Vegetation Index (RECI) successfully differentiated susceptible and tolerant soybean accessions prior to visual symptom development. We report pre-visual detection of soybean wilting using a combination of different vegetation indices. These results can contribute to early stress detection methodologies and rapid classification of drought responses in screening nurseries for breeding and production applications.
How useful is Active Learning for Image-based Plant Phenotyping?
Jul 01, 2020



Abstract:Deep learning models have been successfully deployed for a diverse array of image-based plant phenotyping applications including disease detection and classification. However, successful deployment of supervised deep learning models requires large amount of labeled data, which is a significant challenge in plant science (and most biological) domains due to the inherent complexity. Specifically, data annotation is costly, laborious, time consuming and needs domain expertise for phenotyping tasks, especially for diseases. To overcome this challenge, active learning algorithms have been proposed that reduce the amount of labeling needed by deep learning models to achieve good predictive performance. Active learning methods adaptively select samples to annotate using an acquisition function to achieve maximum (classification) performance under a fixed labeling budget. We report the performance of four different active learning methods, (1) Deep Bayesian Active Learning (DBAL), (2) Entropy, (3) Least Confidence, and (4) Coreset, with conventional random sampling-based annotation for two different image-based classification datasets. The first image dataset consists of soybean [Glycine max L. (Merr.)] leaves belonging to eight different soybean stresses and a healthy class, and the second consists of nine different weed species from the field. For a fixed labeling budget, we observed that the classification performance of deep learning models with active learning-based acquisition strategies is better than random sampling-based acquisition for both datasets. The integration of active learning strategies for data annotation can help mitigate labelling challenges in the plant sciences applications particularly where deep domain knowledge is required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge