Usefulness of interpretability methods to explain deep learning based plant stress phenotyping
Paper and Code
Jul 11, 2020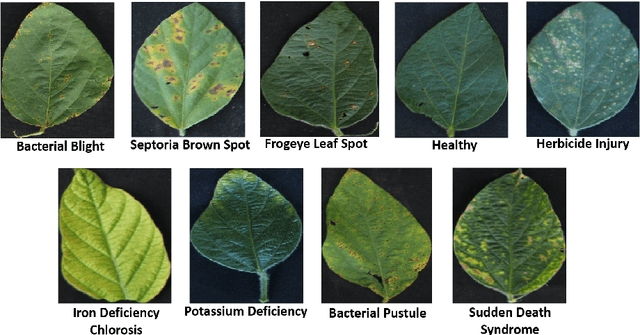



Deep learning techniques have been successfully deployed for automating plant stress identification and quantification. In recent years, there is a growing push towards training models that are interpretable -i.e. that justify their classification decisions by visually highlighting image features that were crucial for classification decisions. The expectation is that trained network models utilize image features that mimic visual cues used by plant pathologists. In this work, we compare some of the most popular interpretability methods: Saliency Maps, SmoothGrad, Guided Backpropogation, Deep Taylor Decomposition, Integrated Gradients, Layer-wise Relevance Propagation and Gradient times Input, for interpreting the deep learning model. We train a DenseNet-121 network for the classification of eight different soybean stresses (biotic and abiotic). Using a dataset consisting of 16,573 RGB images of healthy and stressed soybean leaflets captured under controlled conditions, we obtained an overall classification accuracy of 95.05 \%. For a diverse subset of the test data, we compared the important features with those identified by a human expert. We observed that most interpretability methods identify the infected regions of the leaf as important features for some -- but not all -- of the correctly classified images. For some images, the output of the interpretability methods indicated that spurious feature correlations may have been used to correctly classify them. Although the output explanation maps of these interpretability methods may be different from each other for a given image, we advocate the use of these interpretability methods as `hypothesis generation' mechanisms that can drive scientific insight.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge