Sambuddha Ghosal
Uncertainty Quantified Deep Learning for Predicting Dice Coefficient of Digital Histopathology Image Segmentation
Aug 31, 2021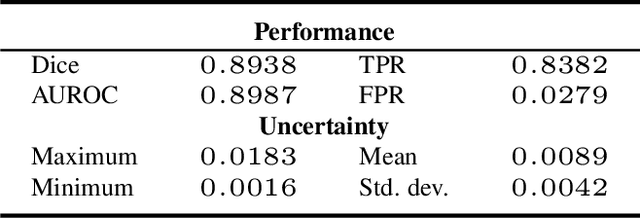
Abstract:Deep learning models (DLMs) can achieve state of the art performance in medical image segmentation and classification tasks. However, DLMs that do not provide feedback for their predictions such as Dice coefficients (Dice) have limited deployment potential in real world clinical settings. Uncertainty estimates can increase the trust of these automated systems by identifying predictions that need further review but remain computationally prohibitive to deploy. In this study, we use a DLM with randomly initialized weights and Monte Carlo dropout (MCD) to segment tumors from microscopic Hematoxylin and Eosin (H&E) dye stained prostate core biopsy RGB images. We devise a novel approach that uses multiple clinical region based uncertainties from a single image (instead of the entire image) to predict Dice of the DLM model output by linear models. Image level uncertainty maps were generated and showed correspondence between imperfect model segmentation and high levels of uncertainty associated with specific prostate tissue regions with or without tumors. Results from this study suggest that linear models can learn coefficients of uncertainty quantified deep learning and correlations ((Spearman's correlation (p<0.05)) to predict Dice scores of specific regions of medical images.
Deep Multi-view Image Fusion for Soybean Yield Estimation in Breeding Applications Deep Multi-view Image Fusion for Soybean Yield Estimation in Breeding Applications
Nov 13, 2020
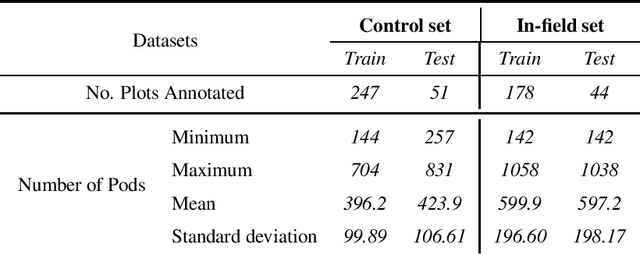
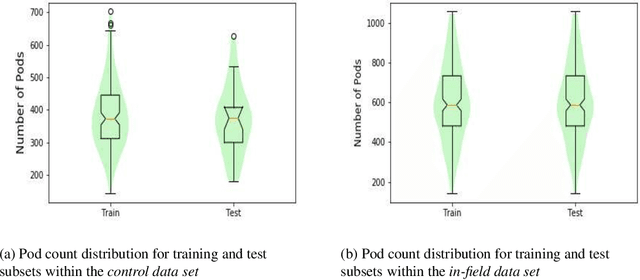
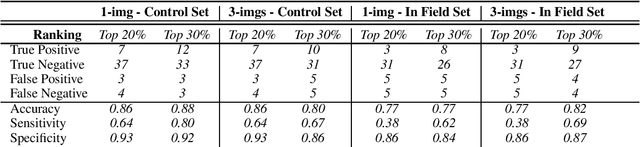
Abstract:Reliable seed yield estimation is an indispensable step in plant breeding programs geared towards cultivar development in major row crops. The objective of this study is to develop a machine learning (ML) approach adept at soybean [\textit{Glycine max} L. (Merr.)] pod counting to enable genotype seed yield rank prediction from in-field video data collected by a ground robot. To meet this goal, we developed a multi-view image-based yield estimation framework utilizing deep learning architectures. Plant images captured from different angles were fused to estimate the yield and subsequently to rank soybean genotypes for application in breeding decisions. We used data from controlled imaging environment in field, as well as from plant breeding test plots in field to demonstrate the efficacy of our framework via comparing performance with manual pod counting and yield estimation. Our results demonstrate the promise of ML models in making breeding decisions with significant reduction of time and human effort, and opening new breeding methods avenues to develop cultivars.
Interpretable and synergistic deep learning for visual explanation and statistical estimations of segmentation of disease features from medical images
Nov 11, 2020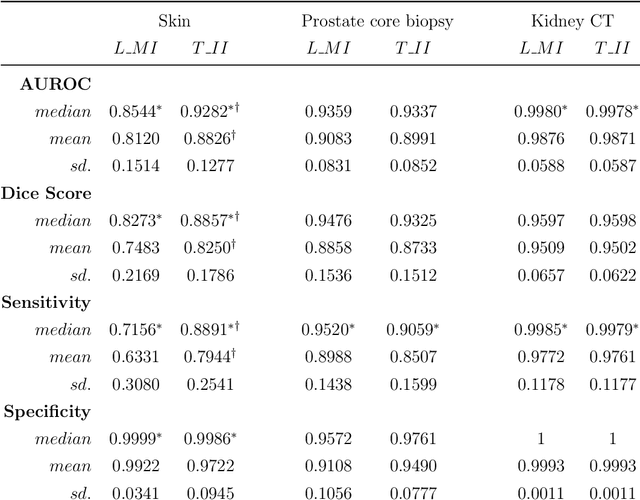
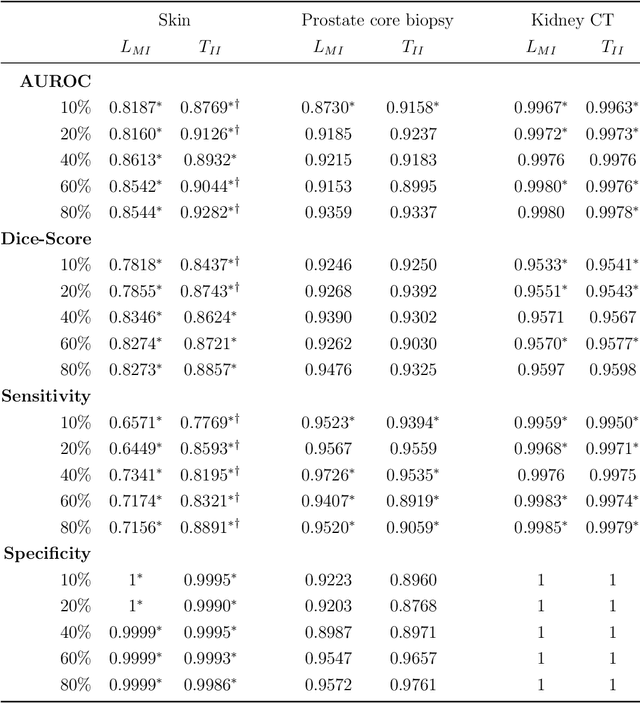
Abstract:Deep learning (DL) models for disease classification or segmentation from medical images are increasingly trained using transfer learning (TL) from unrelated natural world images. However, shortcomings and utility of TL for specialized tasks in the medical imaging domain remain unknown and are based on assumptions that increasing training data will improve performance. We report detailed comparisons, rigorous statistical analysis and comparisons of widely used DL architecture for binary segmentation after TL with ImageNet initialization (TII-models) with supervised learning with only medical images(LMI-models) of macroscopic optical skin cancer, microscopic prostate core biopsy and Computed Tomography (CT) DICOM images. Through visual inspection of TII and LMI model outputs and their Grad-CAM counterparts, our results identify several counter intuitive scenarios where automated segmentation of one tumor by both models or the use of individual segmentation output masks in various combinations from individual models leads to 10% increase in performance. We also report sophisticated ensemble DL strategies for achieving clinical grade medical image segmentation and model explanations under low data regimes. For example; estimating performance, explanations and replicability of LMI and TII models described by us can be used for situations in which sparsity promotes better learning. A free GitHub repository of TII and LMI models, code and more than 10,000 medical images and their Grad-CAM output from this study can be used as starting points for advanced computational medicine and DL research for biomedical discovery and applications.
Deep Generative Models Strike Back! Improving Understanding and Evaluation in Light of Unmet Expectations for OoD Data
Nov 12, 2019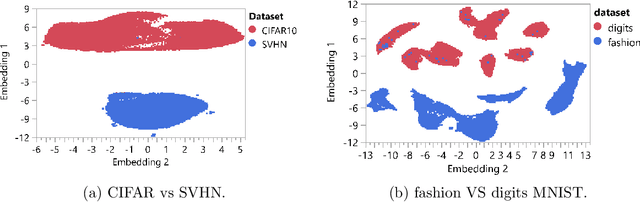
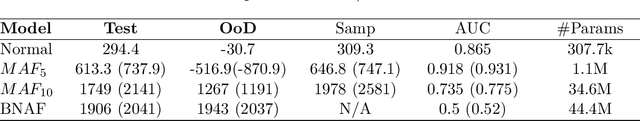

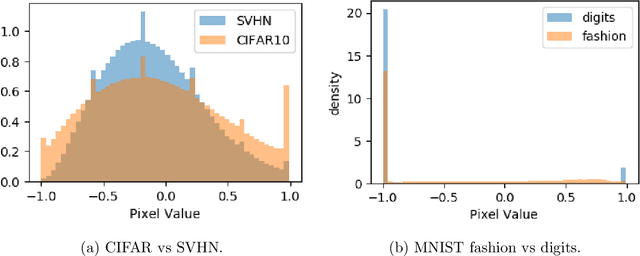
Abstract:Advances in deep generative and density models have shown impressive capacity to model complex probability density functions in lower-dimensional space. Also, applying such models to high-dimensional image data to model the PDF has shown poor generalization, with out-of-distribution data being assigned equal or higher likelihood than in-sample data. Methods to deal with this have been proposed that deviate from a fully unsupervised approach, requiring large ensembles or additional knowledge about the data, not commonly available in the real-world. In this work, the previously offered reasoning behind these issues is challenged empirically, and it is shown that data-sets such as MNIST fashion/digits and CIFAR10/SVHN are trivially separable and have no overlap on their respective data manifolds that explains the higher OoD likelihood. Models like masked autoregressive flows and block neural autoregressive flows are shown to not suffer from OoD likelihood issues to the extent of GLOW, PixelCNN++, and real NVP. A new avenue is also explored which involves a change of basis to a new space of the same dimension with an orthonormal unitary basis of eigenvectors before modeling. In the test data-sets and models, this aids in pushing down the relative likelihood of the contrastive OoD data set and improve discrimination results. The significance of the density of the original space is maintained, while invertibility remains tractable. Finally, a look to the previous generation of generative models in the form of probabilistic principal component analysis is inspired, and revisited for the same data-sets and shown to work really well for discriminating anomalies based on likelihood in a fully unsupervised fashion compared with pixelCNN++, GLOW, and real NVP with less complexity and faster training. Also, dimensionality reduction using PCA is shown to improve anomaly detection in generative models.
Encoding Invariances in Deep Generative Models
Jun 04, 2019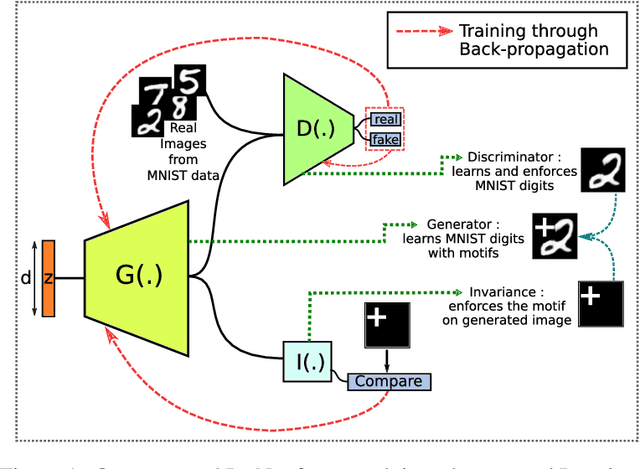
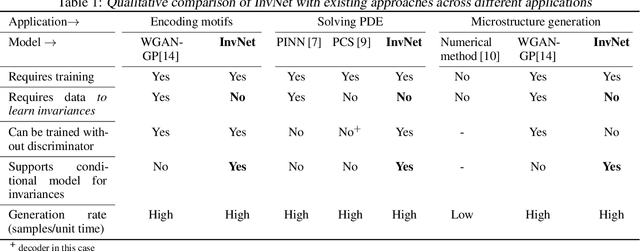
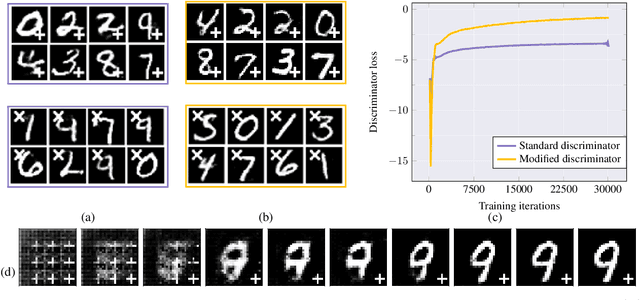
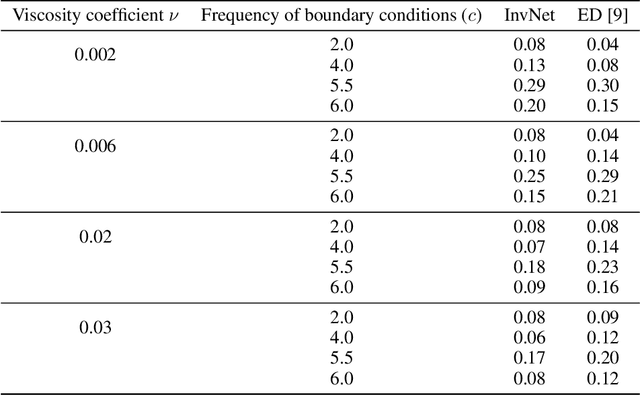
Abstract:Reliable training of generative adversarial networks (GANs) typically require massive datasets in order to model complicated distributions. However, in several applications, training samples obey invariances that are \textit{a priori} known; for example, in complex physics simulations, the training data obey universal laws encoded as well-defined mathematical equations. In this paper, we propose a new generative modeling approach, InvNet, that can efficiently model data spaces with known invariances. We devise an adversarial training algorithm to encode them into data distribution. We validate our framework in three experimental settings: generating images with fixed motifs; solving nonlinear partial differential equations (PDEs); and reconstructing two-phase microstructures with desired statistical properties. We complement our experiments with several theoretical results.
Interpretable deep learning for guided structure-property explorations in photovoltaics
Dec 12, 2018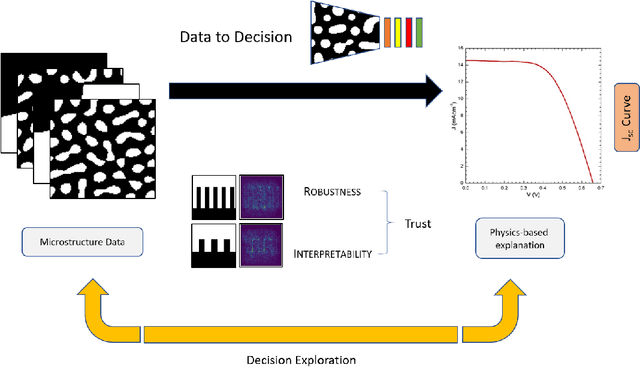
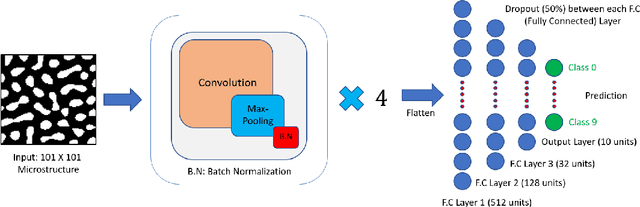
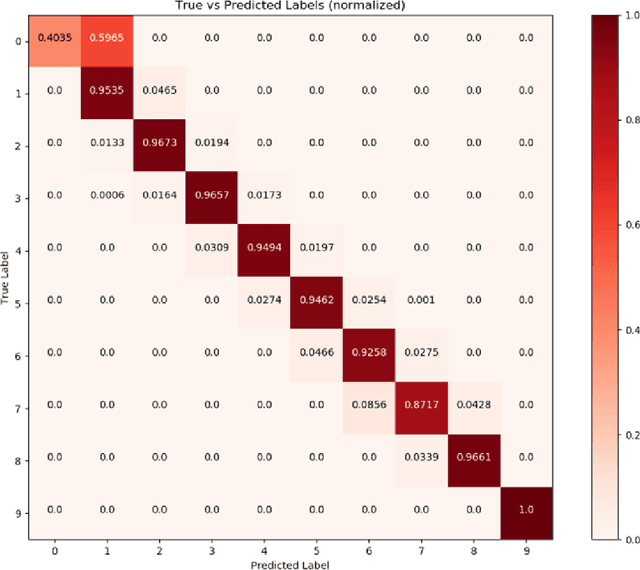
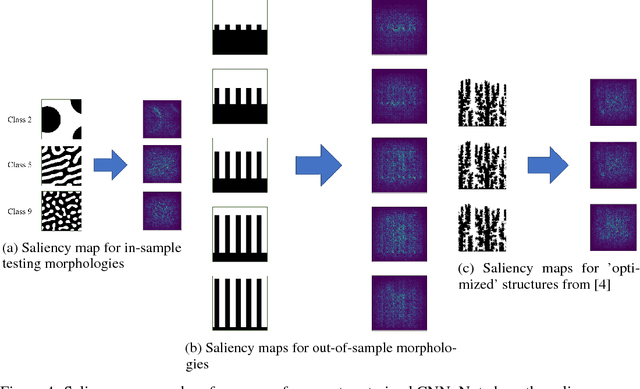
Abstract:The performance of an organic photovoltaic device is intricately connected to its active layer morphology. This connection between the active layer and device performance is very expensive to evaluate, either experimentally or computationally. Hence, designing morphologies to achieve higher performances is non-trivial and often intractable. To solve this, we first introduce a deep convolutional neural network (CNN) architecture that can serve as a fast and robust surrogate for the complex structure-property map. Several tests were performed to gain trust in this trained model. Then, we utilize this fast framework to perform robust microstructural design to enhance device performance.
Interpretable Deep Learning applied to Plant Stress Phenotyping
Oct 28, 2017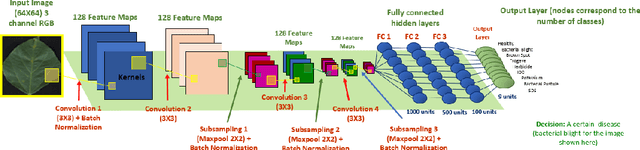
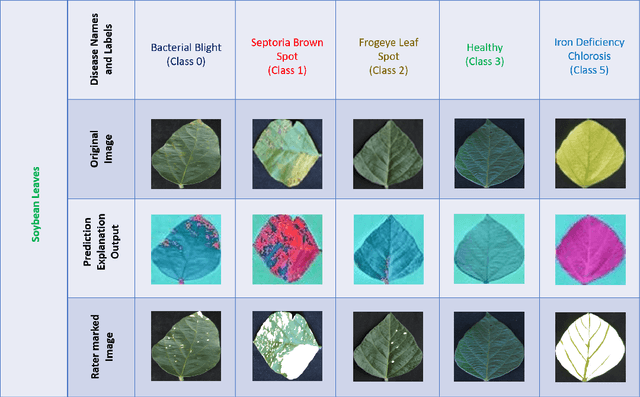
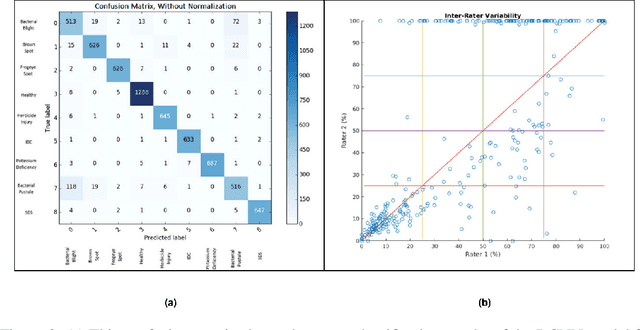
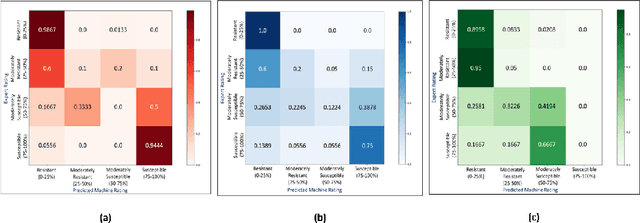
Abstract:Availability of an explainable deep learning model that can be applied to practical real world scenarios and in turn, can consistently, rapidly and accurately identify specific and minute traits in applicable fields of biological sciences, is scarce. Here we consider one such real world example viz., accurate identification, classification and quantification of biotic and abiotic stresses in crop research and production. Up until now, this has been predominantly done manually by visual inspection and require specialized training. However, such techniques are hindered by subjectivity resulting from inter- and intra-rater cognitive variability. Here, we demonstrate the ability of a machine learning framework to identify and classify a diverse set of foliar stresses in the soybean plant with remarkable accuracy. We also present an explanation mechanism using gradient-weighted class activation mapping that isolates the visual symptoms used by the model to make predictions. This unsupervised identification of unique visual symptoms for each stress provides a quantitative measure of stress severity, allowing for identification, classification and quantification in one framework. The learnt model appears to be agnostic to species and make good predictions for other (non-soybean) species, demonstrating an ability of transfer learning.
An unsupervised spatiotemporal graphical modeling approach to anomaly detection in distributed CPS
May 20, 2016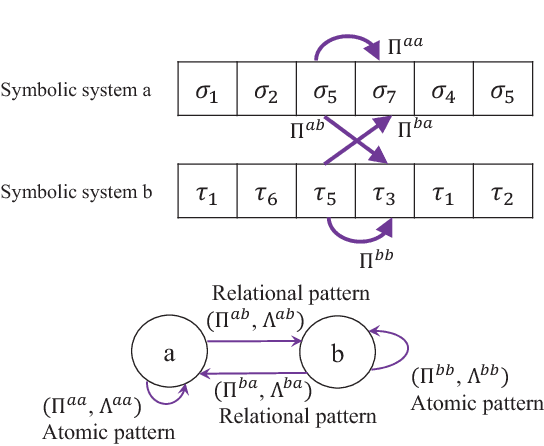
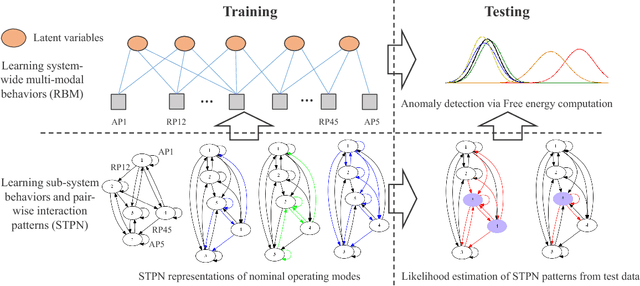
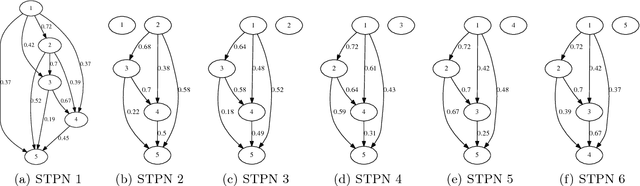
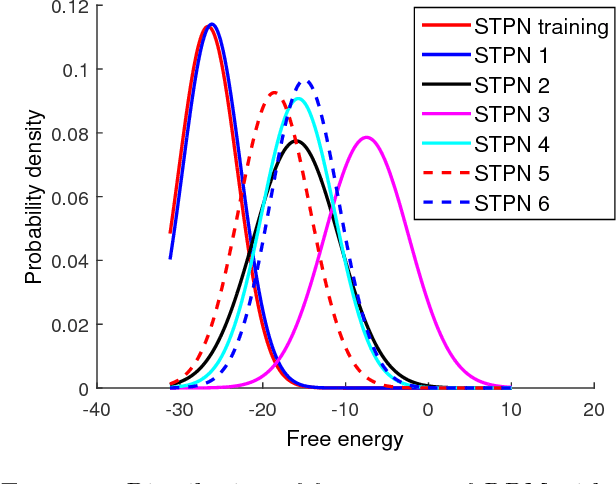
Abstract:Modern distributed cyber-physical systems (CPSs) encounter a large variety of physical faults and cyber anomalies and in many cases, they are vulnerable to catastrophic fault propagation scenarios due to strong connectivity among the sub-systems. This paper presents a new data-driven framework for system-wide anomaly detection for addressing such issues. The framework is based on a spatiotemporal feature extraction scheme built on the concept of symbolic dynamics for discovering and representing causal interactions among the subsystems of a CPS. The extracted spatiotemporal features are then used to learn system-wide patterns via a Restricted Boltzmann Machine (RBM). The results show that: (1) the RBM free energy in the off-nominal conditions is different from that in the nominal conditions and can be used for anomaly detection; (2) the framework can capture multiple nominal modes with one graphical model; (3) the case studies with simulated data and an integrated building system validate the proposed approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge