Pratik Shah
Uncertainty Quantified Deep Learning and Regression Analysis Framework for Image Segmentation of Skin Cancer Lesions
Dec 28, 2024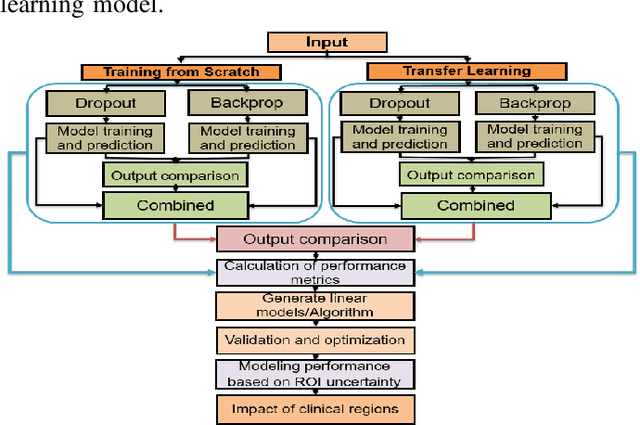
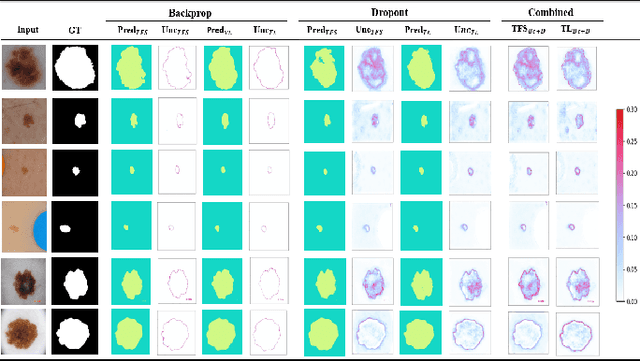
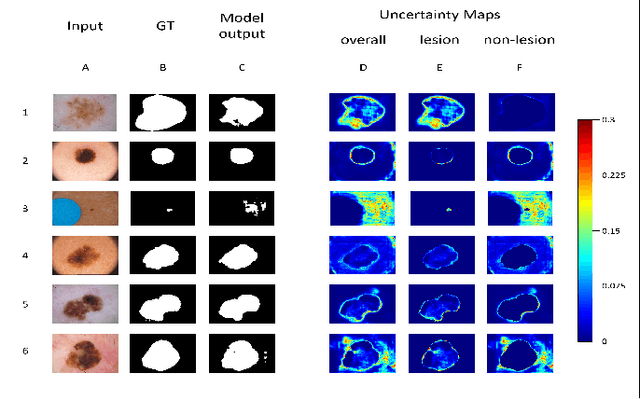
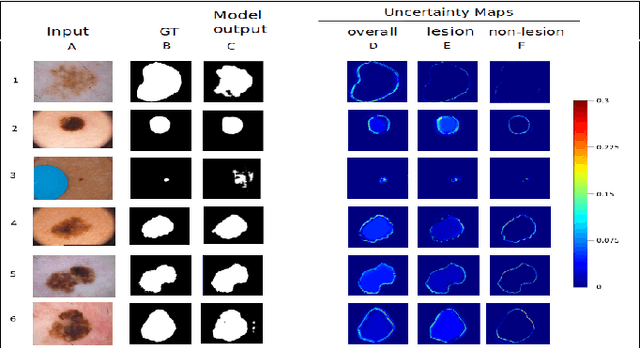
Abstract:Deep learning models (DLMs) frequently achieve accurate segmentation and classification of tumors from medical images. However, DLMs lacking feedback on their image segmentation mechanisms, such as Dice coefficients and confidence in their performance, face challenges when processing previously unseen images in real-world clinical settings. Uncertainty estimates to identify DLM predictions at the cellular or single-pixel level that require clinician review can enhance trust. However, their deployment requires significant computational resources. This study reports two DLMs, one trained from scratch and another based on transfer learning, with Monte Carlo dropout or Bayes-by-backprop uncertainty estimations to segment lesions from the publicly available The International Skin Imaging Collaboration-19 dermoscopy image database with cancerous lesions. A novel approach to compute pixel-by-pixel uncertainty estimations of DLM segmentation performance in multiple clinical regions from a single dermoscopy image with corresponding Dice scores is reported for the first time. Image-level uncertainty maps demonstrated correspondence between imperfect DLM segmentation and high uncertainty levels in specific skin tissue regions, with or without lesions. Four new linear regression models that can predict the Dice performance of DLM segmentation using constants and uncertainty measures, either individually or in combination from lesions, tissue structures, and non-tissue pixel regions critical for clinical diagnosis and prognostication in skin images (Spearman's correlation, p < 0.05), are reported for the first time for low-compute uncertainty estimation workflows.
Lagrangian Index Policy for Restless Bandits with Average Reward
Dec 17, 2024



Abstract:We study the Lagrangian Index Policy (LIP) for restless multi-armed bandits with long-run average reward. In particular, we compare the performance of LIP with the performance of the Whittle Index Policy (WIP), both heuristic policies known to be asymptotically optimal under certain natural conditions. Even though in most cases their performances are very similar, in the cases when WIP shows bad performance, LIP continues to perform very well. We then propose reinforcement learning algorithms, both tabular and NN-based, to obtain online learning schemes for LIP in the model-free setting. The proposed reinforcement learning schemes for LIP requires significantly less memory than the analogous scheme for WIP. We calculate analytically the Lagrangian index for the restart model, which describes the optimal web crawling and the minimization of the weighted age of information. We also give a new proof of asymptotic optimality in case of homogeneous bandits as the number of arms goes to infinity, based on exchangeability and de Finetti's theorem.
Responsible Deep Learning for Software as a Medical Device
Dec 20, 2023Abstract:Tools, models and statistical methods for signal processing and medical image analysis and training deep learning models to create research prototypes for eventual clinical applications are of special interest to the biomedical imaging community. But material and optical properties of biological tissues are complex and not easily captured by imaging devices. Added complexity can be introduced by datasets with underrepresentation of medical images from races and ethnicities for deep learning, and limited knowledge about the regulatory framework needed for commercialization and safety of emerging Artificial Intelligence (AI) and Machine Learning (ML) technologies for medical image analysis. This extended version of the workshop paper presented at the special session of the 2022 IEEE 19th International Symposium on Biomedical Imaging, describes strategy and opportunities by University of California professors engaged in machine learning (section I) and clinical research (section II), the Office of Science and Engineering Laboratories (OSEL) section III, and officials at the US FDA in Center for Devices & Radiological Health (CDRH) section IV. Performance evaluations of AI/ML models of skin (RGB), tissue biopsy (digital pathology), and lungs and kidneys (Magnetic Resonance, X-ray, Computed Tomography) medical images for regulatory evaluations and real-world deployment are discussed.
Uncertainty Quantified Deep Learning for Predicting Dice Coefficient of Digital Histopathology Image Segmentation
Aug 31, 2021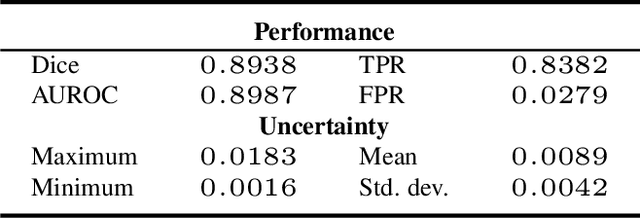
Abstract:Deep learning models (DLMs) can achieve state of the art performance in medical image segmentation and classification tasks. However, DLMs that do not provide feedback for their predictions such as Dice coefficients (Dice) have limited deployment potential in real world clinical settings. Uncertainty estimates can increase the trust of these automated systems by identifying predictions that need further review but remain computationally prohibitive to deploy. In this study, we use a DLM with randomly initialized weights and Monte Carlo dropout (MCD) to segment tumors from microscopic Hematoxylin and Eosin (H&E) dye stained prostate core biopsy RGB images. We devise a novel approach that uses multiple clinical region based uncertainties from a single image (instead of the entire image) to predict Dice of the DLM model output by linear models. Image level uncertainty maps were generated and showed correspondence between imperfect model segmentation and high levels of uncertainty associated with specific prostate tissue regions with or without tumors. Results from this study suggest that linear models can learn coefficients of uncertainty quantified deep learning and correlations ((Spearman's correlation (p<0.05)) to predict Dice scores of specific regions of medical images.
Interpretable and synergistic deep learning for visual explanation and statistical estimations of segmentation of disease features from medical images
Nov 11, 2020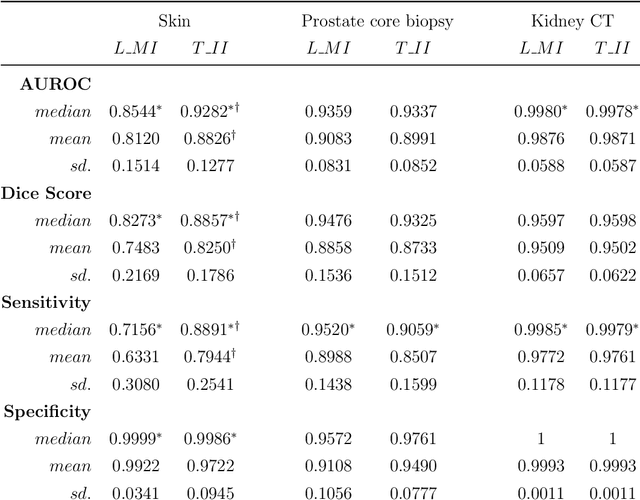
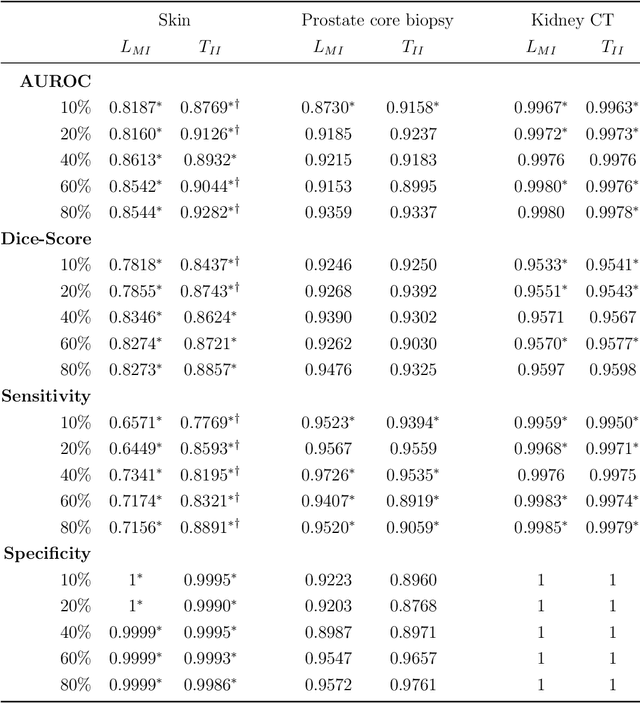
Abstract:Deep learning (DL) models for disease classification or segmentation from medical images are increasingly trained using transfer learning (TL) from unrelated natural world images. However, shortcomings and utility of TL for specialized tasks in the medical imaging domain remain unknown and are based on assumptions that increasing training data will improve performance. We report detailed comparisons, rigorous statistical analysis and comparisons of widely used DL architecture for binary segmentation after TL with ImageNet initialization (TII-models) with supervised learning with only medical images(LMI-models) of macroscopic optical skin cancer, microscopic prostate core biopsy and Computed Tomography (CT) DICOM images. Through visual inspection of TII and LMI model outputs and their Grad-CAM counterparts, our results identify several counter intuitive scenarios where automated segmentation of one tumor by both models or the use of individual segmentation output masks in various combinations from individual models leads to 10% increase in performance. We also report sophisticated ensemble DL strategies for achieving clinical grade medical image segmentation and model explanations under low data regimes. For example; estimating performance, explanations and replicability of LMI and TII models described by us can be used for situations in which sparsity promotes better learning. A free GitHub repository of TII and LMI models, code and more than 10,000 medical images and their Grad-CAM output from this study can be used as starting points for advanced computational medicine and DL research for biomedical discovery and applications.
High Accuracy Tumor Diagnoses and Benchmarking of Hematoxylin and Eosin Stained Prostate Core Biopsy Images Generated by Explainable Deep Neural Networks
Aug 02, 2019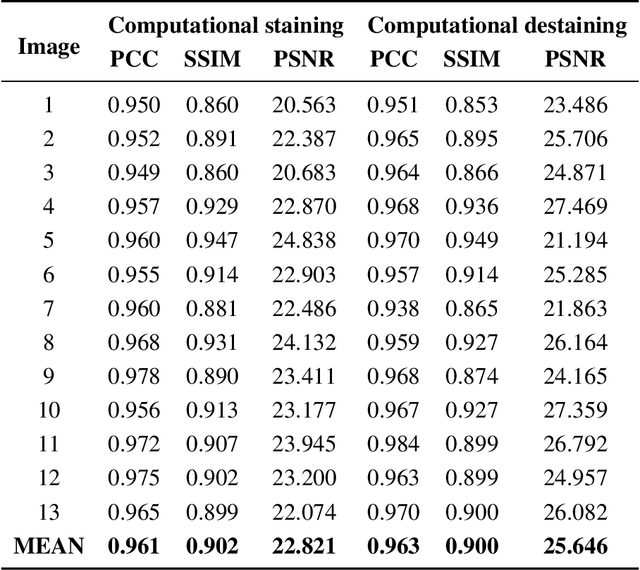
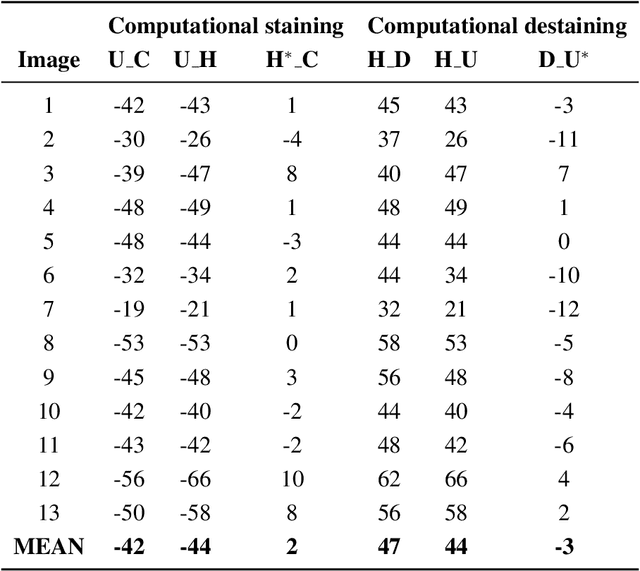
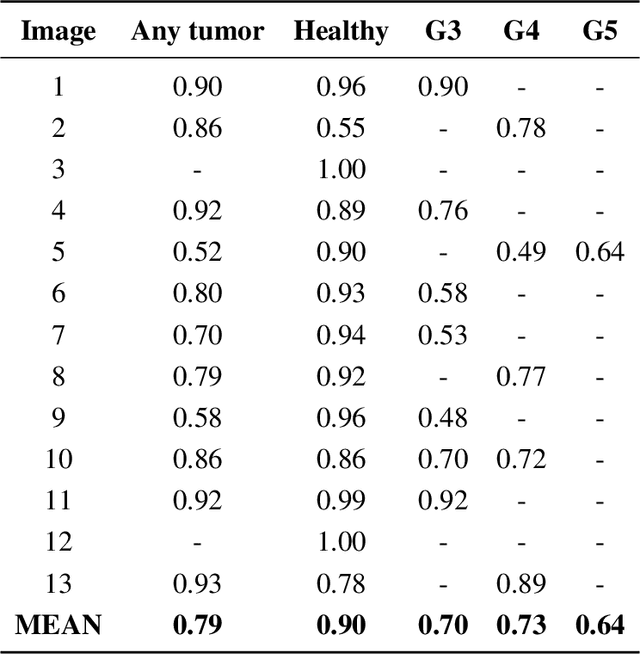
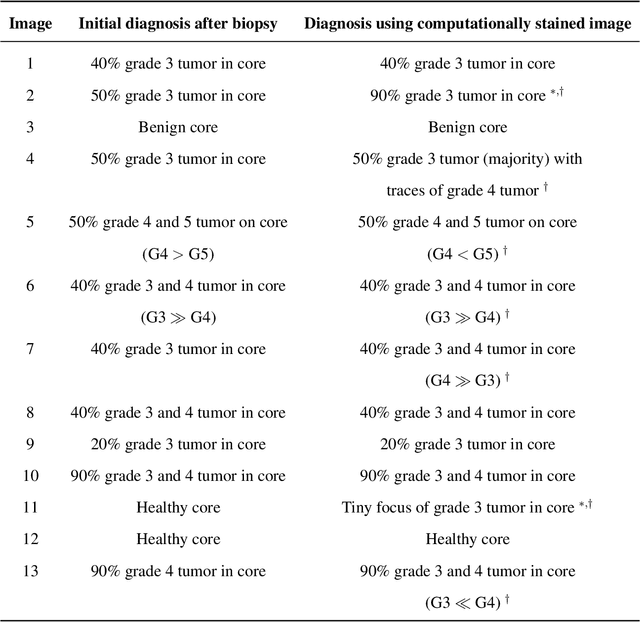
Abstract:Histopathological diagnoses of tumors in tissue biopsy after Hematoxylin and Eosin (H&E) staining is the gold standard for oncology care. H&E staining is slow and uses dyes, reagents and precious tissue samples that cannot be reused. Thousands of native nonstained RGB Whole Slide Image (RWSI) patches of prostate core tissue biopsies were registered with their H&E stained versions. Conditional Generative Adversarial Neural Networks (cGANs) that automate conversion of native nonstained RWSI to computational H&E stained images were then trained. High similarities between computational and H&E dye stained images with Structural Similarity Index (SSIM) 0.902, Pearsons Correlation Coefficient (CC) 0.962 and Peak Signal to Noise Ratio (PSNR) 22.821 dB were calculated. A second cGAN performed accurate computational destaining of H&E dye stained images back to their native nonstained form with SSIM 0.9, CC 0.963 and PSNR 25.646 dB. A single-blind study computed more than 95% pixel-by-pixel overlap between prostate tumor annotations on computationally stained images, provided by five-board certified MD pathologists, with those on H&E dye stained counterparts. We report the first visualization and explanation of neural network kernel activation maps during H&E staining and destaining of RGB images by cGANs. High similarities between kernel activation maps of computational and H&E stained images (Mean-Squared Errors <0.0005) provide additional mathematical and mechanistic validation of the staining system. Our neural network framework thus is automated, explainable and performs high precision H&E staining and destaining of low cost native RGB images, and is computer vision and physician authenticated for rapid and accurate tumor diagnoses.
Automated Process Incorporating Machine Learning Segmentation and Correlation of Oral Diseases with Systemic Health
Oct 25, 2018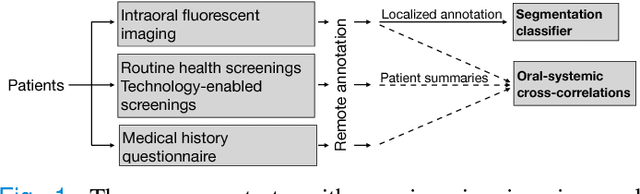

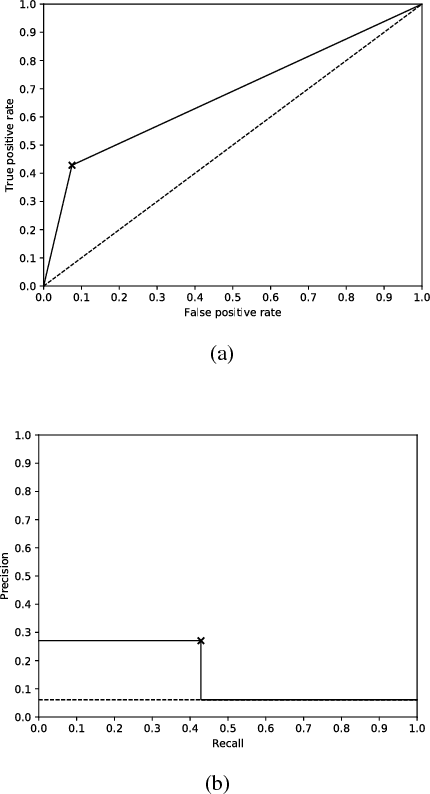

Abstract:Imaging fluorescent disease biomarkers in tissues and skin is a non-invasive method to screen for health conditions. We report an automated process that combines intraoral fluorescent porphyrin biomarker imaging, clinical examinations and machine learning for correlation of systemic health conditions with periodontal disease. 1215 intraoral fluorescent images, from 284 consenting adults aged 18-90, were analyzed using a machine learning classifier that can segment periodontal inflammation. The classifier achieved an AUC of 0.677 with precision and recall of 0.271 and 0.429, respectively, indicating a learned association between disease signatures in collected images. Periodontal diseases were more prevalent among males (p=0.0012) and older subjects (p=0.0224) in the screened population. Physicians independently examined the collected images, assigning localized modified gingival indices (MGIs). MGIs and periodontal disease were then cross-correlated with responses to a medical history questionnaire, blood pressure and body mass index measurements, and optic nerve, tympanic membrane, neurological, and cardiac rhythm imaging examinations. Gingivitis and early periodontal disease were associated with subjects diagnosed with optic nerve abnormalities (p <0.0001) in their retinal scans. We also report significant co-occurrences of periodontal disease in subjects reporting swollen joints (p=0.0422) and a family history of eye disease (p=0.0337). These results indicate cross-correlation of poor periodontal health with systemic health outcomes and stress the importance of oral health screenings at the primary care level. Our screening process and analysis method, using images and machine learning, can be generalized for automated diagnoses and systemic health screenings for other diseases.
Curve Reconstruction in Riemannian Manifolds: Ordering Motion Frames
Dec 20, 2010



Abstract:In this article we extend the computational geometric curve reconstruction approach to curves in Riemannian manifolds. We prove that the minimal spanning tree, given a sufficiently dense sample, correctly reconstructs the smooth arcs and further closed and simple curves in Riemannian manifolds. The proof is based on the behaviour of the curve segment inside the tubular neighbourhood of the curve. To take care of the local topological changes of the manifold, the tubular neighbourhood is constructed in consideration with the injectivity radius of the underlying Riemannian manifold. We also present examples of successfully reconstructed curves and show an applications of curve reconstruction to ordering motion frames.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge