Uncertainty Quantified Deep Learning and Regression Analysis Framework for Image Segmentation of Skin Cancer Lesions
Paper and Code
Dec 28, 2024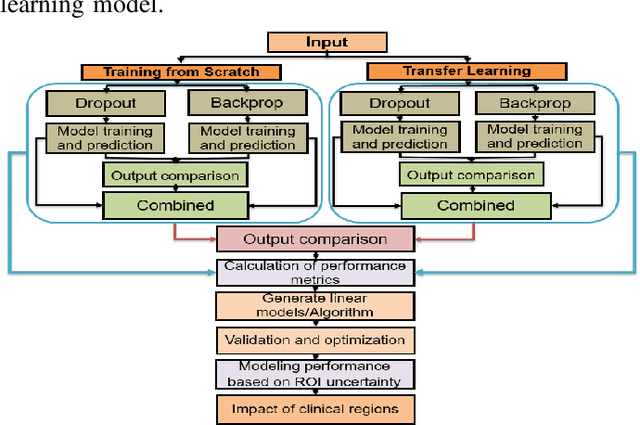
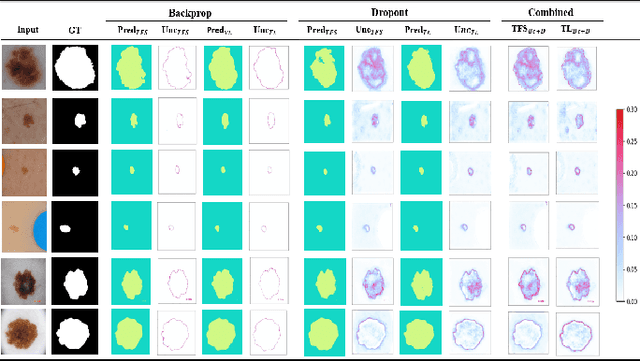
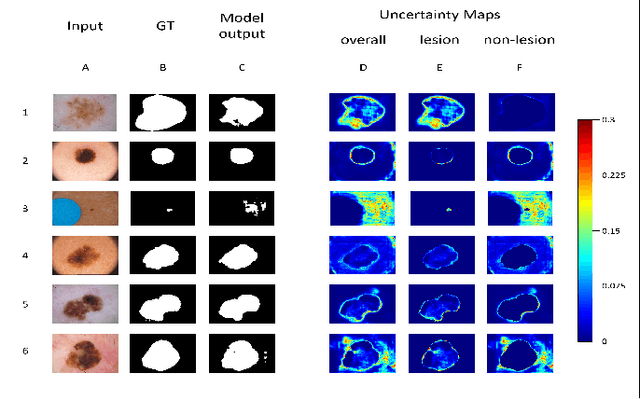
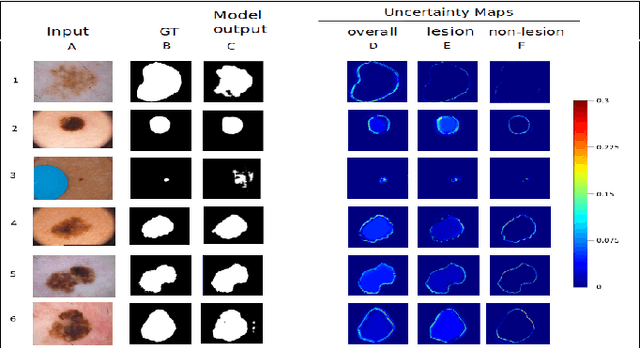
Deep learning models (DLMs) frequently achieve accurate segmentation and classification of tumors from medical images. However, DLMs lacking feedback on their image segmentation mechanisms, such as Dice coefficients and confidence in their performance, face challenges when processing previously unseen images in real-world clinical settings. Uncertainty estimates to identify DLM predictions at the cellular or single-pixel level that require clinician review can enhance trust. However, their deployment requires significant computational resources. This study reports two DLMs, one trained from scratch and another based on transfer learning, with Monte Carlo dropout or Bayes-by-backprop uncertainty estimations to segment lesions from the publicly available The International Skin Imaging Collaboration-19 dermoscopy image database with cancerous lesions. A novel approach to compute pixel-by-pixel uncertainty estimations of DLM segmentation performance in multiple clinical regions from a single dermoscopy image with corresponding Dice scores is reported for the first time. Image-level uncertainty maps demonstrated correspondence between imperfect DLM segmentation and high uncertainty levels in specific skin tissue regions, with or without lesions. Four new linear regression models that can predict the Dice performance of DLM segmentation using constants and uncertainty measures, either individually or in combination from lesions, tissue structures, and non-tissue pixel regions critical for clinical diagnosis and prognostication in skin images (Spearman's correlation, p < 0.05), are reported for the first time for low-compute uncertainty estimation workflows.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge