Aneta Lisowska
Defining Effective Engagement For Enhancing Cancer Patients' Well-being with Mobile Digital Behavior Change Interventions
Mar 19, 2024



Abstract:Digital Behavior Change Interventions (DBCIs) are supporting development of new health behaviors. Evaluating their effectiveness is crucial for their improvement and understanding of success factors. However, comprehensive guidance for developers, particularly in small-scale studies with ethical constraints, is limited. Building on the CAPABLE project, this study aims to define effective engagement with DBCIs for supporting cancer patients in enhancing their quality of life. We identify metrics for measuring engagement, explore the interest of both patients and clinicians in DBCIs, and propose hypotheses for assessing the impact of DBCIs in such contexts. Our findings suggest that clinician prescriptions significantly increase sustained engagement with mobile DBCIs. In addition, while one weekly engagement with a DBCI is sufficient to maintain well-being, transitioning from extrinsic to intrinsic motivation may require a higher level of engagement.
Decoding Emotional Valence from Wearables: Can Our Data Reveal Our True Feelings?
Dec 21, 2023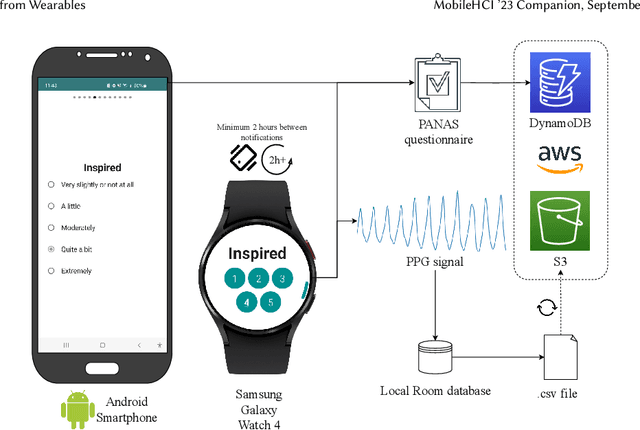
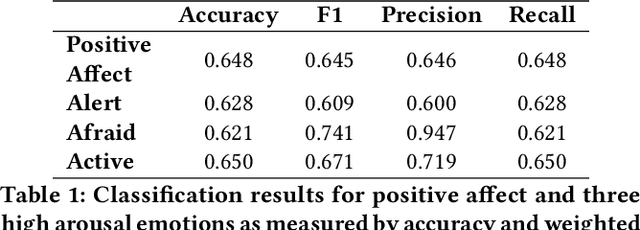
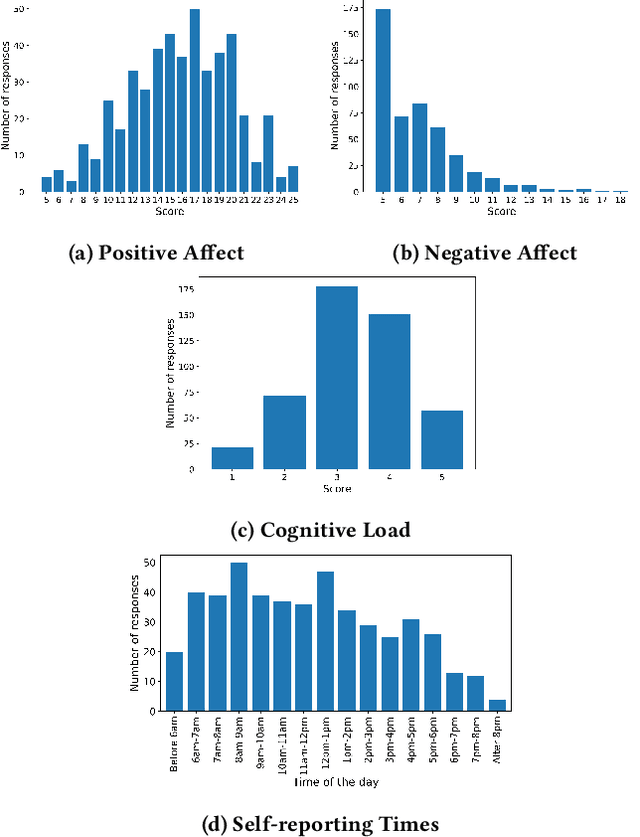
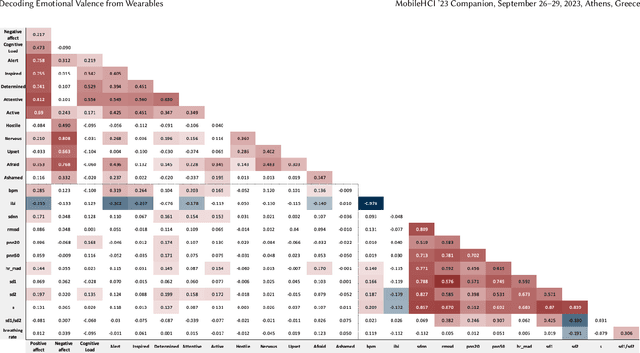
Abstract:Automatic detection and tracking of emotional states has the potential for helping individuals with various mental health conditions. While previous studies have captured physiological signals using wearable devices in laboratory settings, providing valuable insights into the relationship between physiological responses and mental states, the transfer of these findings to real-life scenarios is still in its nascent stages. Our research aims to bridge the gap between laboratory-based studies and real-life settings by leveraging consumer-grade wearables and self-report measures. We conducted a preliminary study involving 15 healthy participants to assess the efficacy of wearables in capturing user valence in real-world settings. In this paper, we present the initial analysis of the collected data, focusing primarily on the results of valence classification. Our findings demonstrate promising results in distinguishing between high and low positive valence, achieving an F1 score of 0.65. This research opens up avenues for future research in the field of mobile mental health interventions.
Personalizing Digital Health Behavior Change Interventions using Machine Learning and Domain Knowledge
Apr 14, 2023Abstract:We are developing a virtual coaching system that helps patients adhere to behavior change interventions (BCI). Our proposed system predicts whether a patient will perform the targeted behavior and uses counterfactual examples with feature control to guide personalizsation of BCI. We evaluated our prediction model using simulated patient data with varying levels of receptivity to intervention.
Can gamification reduce the burden of self-reporting in mHealth applications? Feasibility study using machine learning from smartwatch data to estimate cognitive load
Feb 07, 2023Abstract:The effectiveness of digital treatments can be measured by requiring patients to self-report their mental and physical state through mobile applications. However, self-reporting can be overwhelming and may cause patients to disengage from the intervention. In order to address this issue, we conduct a feasibility study to explore the impact of gamification on the cognitive burden of self-reporting. Our approach involves the creation of a system to assess cognitive burden through the analysis of photoplethysmography (PPG) signals obtained from a smartwatch. The system is built by collecting PPG data during both cognitively demanding tasks and periods of rest. The obtained data is utilized to train a machine learning model to detect cognitive load (CL). Subsequently, we create two versions of health surveys: a gamified version and a traditional version. Our aim is to estimate the cognitive load experienced by participants while completing these surveys using their mobile devices. We find that CL detector performance can be enhanced via pre-training on stress detection tasks and requires capturing of a minimum 30 seconds of PPG signal to work adequately. For 10 out of 13 participants, a personalized cognitive load detector can achieve an F1 score above 0.7. We find no difference between the gamified and non-gamified mobile surveys in terms of time spent in the state of high cognitive load but participants prefer the gamified version. The average time spent on each question is 5.5 for gamified survey vs 6 seconds for the non-gamified version.
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022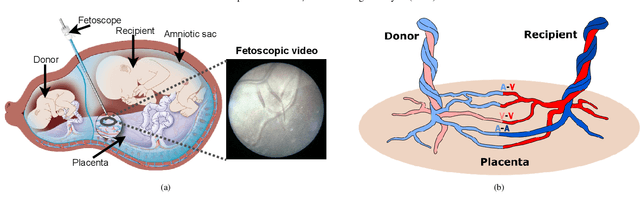
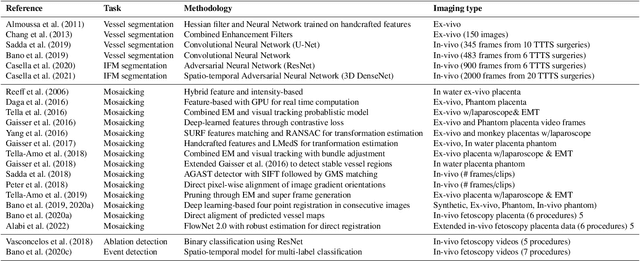
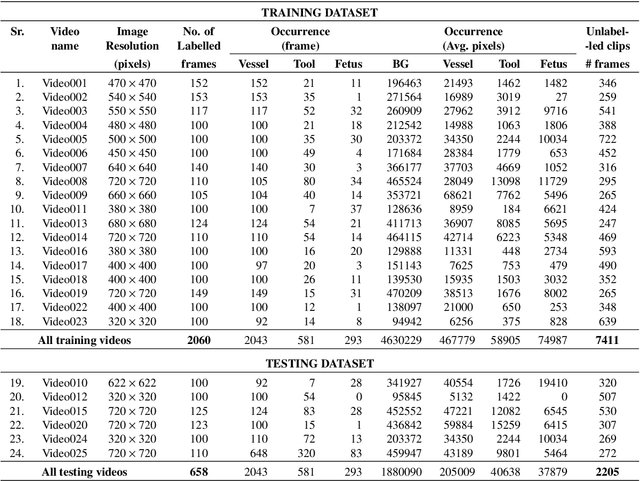
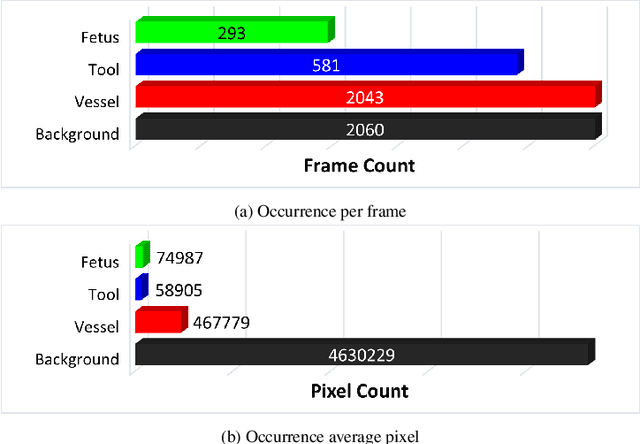
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
Deep Learning Fetal Ultrasound Video Model Match Human Observers in Biometric Measurements
May 27, 2022Abstract:Objective. This work investigates the use of deep convolutional neural networks (CNN) to automatically perform measurements of fetal body parts, including head circumference, biparietal diameter, abdominal circumference and femur length, and to estimate gestational age and fetal weight using fetal ultrasound videos. Approach. We developed a novel multi-task CNN-based spatio-temporal fetal US feature extraction and standard plane detection algorithm (called FUVAI) and evaluated the method on 50 freehand fetal US video scans. We compared FUVAI fetal biometric measurements with measurements made by five experienced sonographers at two time points separated by at least two weeks. Intra- and inter-observer variabilities were estimated. Main results. We found that automated fetal biometric measurements obtained by FUVAI were comparable to the measurements performed by experienced sonographers The observed differences in measurement values were within the range of inter- and intra-observer variability. Moreover, analysis has shown that these differences were not statistically significant when comparing any individual medical expert to our model. Significance. We argue that FUVAI has the potential to assist sonographers who perform fetal biometric measurements in clinical settings by providing them with suggestions regarding the best measuring frames, along with automated measurements. Moreover, FUVAI is able perform these tasks in just a few seconds, which is a huge difference compared to the average of six minutes taken by sonographers. This is significant, given the shortage of medical experts capable of interpreting fetal ultrasound images in numerous countries.
* Published at Physics in Medicine & Biology
CXR-FL: Deep Learning-based Chest X-ray Image Analysis Using Federated Learning
Apr 11, 2022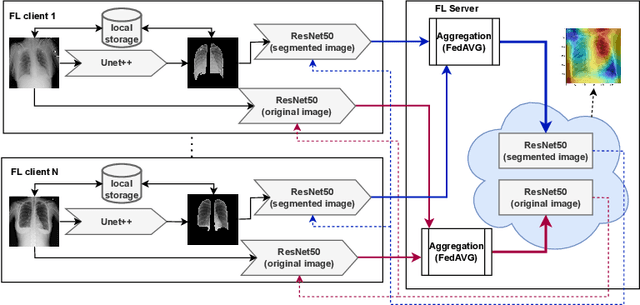
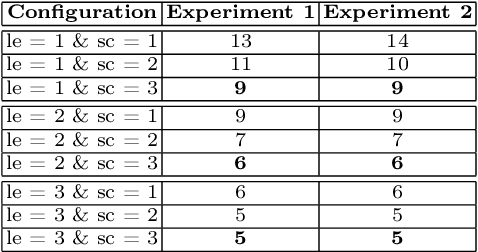
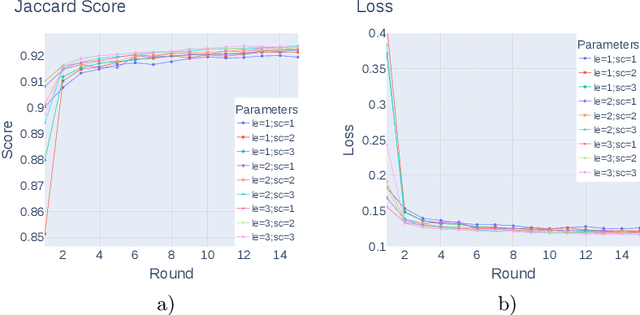
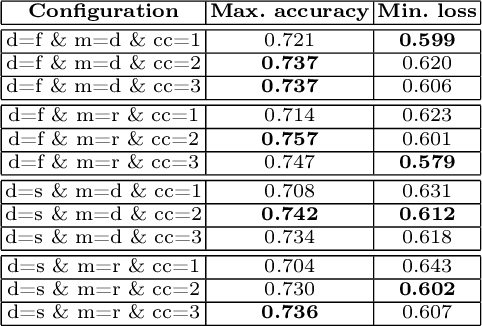
Abstract:Federated learning enables building a shared model from multicentre data while storing the training data locally for privacy. In this paper, we present an evaluation (called CXR-FL) of deep learning-based models for chest X-ray image analysis using the federated learning method. We examine the impact of federated learning parameters on the performance of central models. Additionally, we show that classification models perform worse if trained on a region of interest reduced to segmentation of the lung compared to the full image. However, focusing training of the classification model on the lung area may result in improved pathology interpretability during inference. We also find that federated learning helps maintain model generalizability. The pre-trained weights and code are publicly available at (https://github.com/SanoScience/CXR-FL).
Continual Class Incremental Learning for CT Thoracic Segmentation
Aug 12, 2020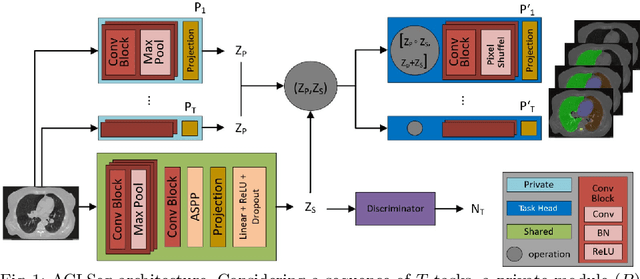

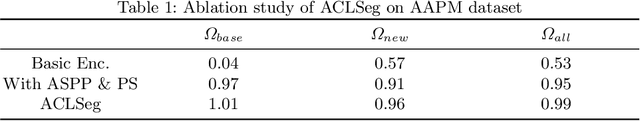
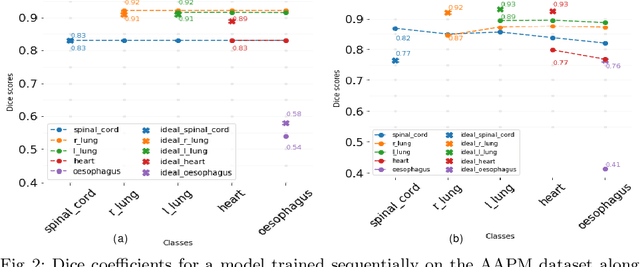
Abstract:Deep learning organ segmentation approaches require large amounts of annotated training data, which is limited in supply due to reasons of confidentiality and the time required for expert manual annotation. Therefore, being able to train models incrementally without having access to previously used data is desirable. A common form of sequential training is fine tuning (FT). In this setting, a model learns a new task effectively, but loses performance on previously learned tasks. The Learning without Forgetting (LwF) approach addresses this issue via replaying its own prediction for past tasks during model training. In this work, we evaluate FT and LwF for class incremental learning in multi-organ segmentation using the publicly available AAPM dataset. We show that LwF can successfully retain knowledge on previous segmentations, however, its ability to learn a new class decreases with the addition of each class. To address this problem we propose an adversarial continual learning segmentation approach (ACLSeg), which disentangles feature space into task-specific and task-invariant features. This enables preservation of performance on past tasks and effective acquisition of new knowledge.
Paying Per-label Attention for Multi-label Extraction from Radiology Reports
Aug 07, 2020Abstract:Training medical image analysis models requires large amounts of expertly annotated data which is time-consuming and expensive to obtain. Images are often accompanied by free-text radiology reports which are a rich source of information. In this paper, we tackle the automated extraction of structured labels from head CT reports for imaging of suspected stroke patients, using deep learning. Firstly, we propose a set of 31 labels which correspond to radiographic findings (e.g. hyperdensity) and clinical impressions (e.g. haemorrhage) related to neurological abnormalities. Secondly, inspired by previous work, we extend existing state-of-the-art neural network models with a label-dependent attention mechanism. Using this mechanism and simple synthetic data augmentation, we are able to robustly extract many labels with a single model, classified according to the radiologist's reporting (positive, uncertain, negative). This approach can be used in further research to effectively extract many labels from medical text.
Teacher-Student chain for efficient semi-supervised histology image classification
Mar 20, 2020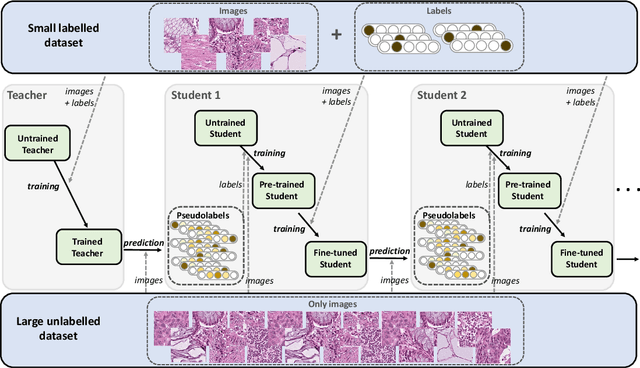

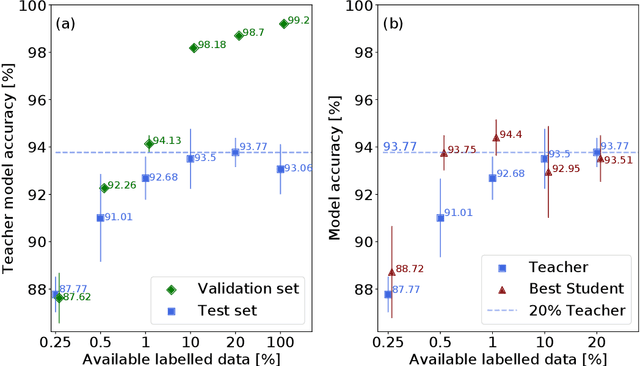
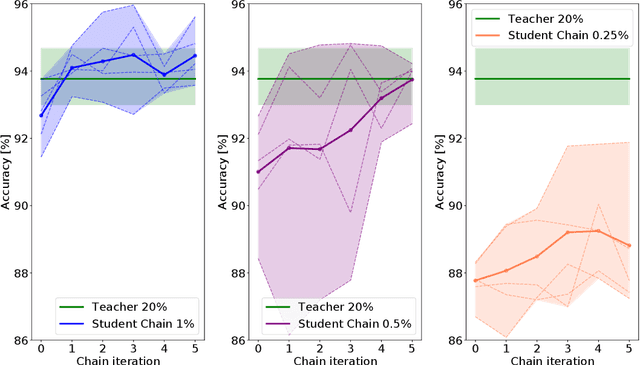
Abstract:Deep learning shows great potential for the domain of digital pathology. An automated digital pathology system could serve as a second reader, perform initial triage in large screening studies, or assist in reporting. However, it is expensive to exhaustively annotate large histology image databases, since medical specialists are a scarce resource. In this paper, we apply the semi-supervised teacher-student knowledge distillation technique proposed by Yalniz et al. (2019) to the task of quantifying prognostic features in colorectal cancer. We obtain accuracy improvements through extending this approach to a chain of students, where each student's predictions are used to train the next student i.e. the student becomes the teacher. Using the chain approach, and only 0.5% labelled data (the remaining 99.5% in the unlabelled pool), we match the accuracy of training on 100% labelled data. At lower percentages of labelled data, similar gains in accuracy are seen, allowing some recovery of accuracy even from a poor initial choice of labelled training set. In conclusion, this approach shows promise for reducing the annotation burden, thus increasing the affordability of automated digital pathology systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge