Michał Lipa
TabAttention: Learning Attention Conditionally on Tabular Data
Oct 27, 2023Abstract:Medical data analysis often combines both imaging and tabular data processing using machine learning algorithms. While previous studies have investigated the impact of attention mechanisms on deep learning models, few have explored integrating attention modules and tabular data. In this paper, we introduce TabAttention, a novel module that enhances the performance of Convolutional Neural Networks (CNNs) with an attention mechanism that is trained conditionally on tabular data. Specifically, we extend the Convolutional Block Attention Module to 3D by adding a Temporal Attention Module that uses multi-head self-attention to learn attention maps. Furthermore, we enhance all attention modules by integrating tabular data embeddings. Our approach is demonstrated on the fetal birth weight (FBW) estimation task, using 92 fetal abdominal ultrasound video scans and fetal biometry measurements. Our results indicate that TabAttention outperforms clinicians and existing methods that rely on tabular and/or imaging data for FBW prediction. This novel approach has the potential to improve computer-aided diagnosis in various clinical workflows where imaging and tabular data are combined. We provide a source code for integrating TabAttention in CNNs at https://github.com/SanoScience/Tab-Attention.
Deep Learning Fetal Ultrasound Video Model Match Human Observers in Biometric Measurements
May 27, 2022Abstract:Objective. This work investigates the use of deep convolutional neural networks (CNN) to automatically perform measurements of fetal body parts, including head circumference, biparietal diameter, abdominal circumference and femur length, and to estimate gestational age and fetal weight using fetal ultrasound videos. Approach. We developed a novel multi-task CNN-based spatio-temporal fetal US feature extraction and standard plane detection algorithm (called FUVAI) and evaluated the method on 50 freehand fetal US video scans. We compared FUVAI fetal biometric measurements with measurements made by five experienced sonographers at two time points separated by at least two weeks. Intra- and inter-observer variabilities were estimated. Main results. We found that automated fetal biometric measurements obtained by FUVAI were comparable to the measurements performed by experienced sonographers The observed differences in measurement values were within the range of inter- and intra-observer variability. Moreover, analysis has shown that these differences were not statistically significant when comparing any individual medical expert to our model. Significance. We argue that FUVAI has the potential to assist sonographers who perform fetal biometric measurements in clinical settings by providing them with suggestions regarding the best measuring frames, along with automated measurements. Moreover, FUVAI is able perform these tasks in just a few seconds, which is a huge difference compared to the average of six minutes taken by sonographers. This is significant, given the shortage of medical experts capable of interpreting fetal ultrasound images in numerous countries.
* Published at Physics in Medicine & Biology
BabyNet: Residual Transformer Module for Birth Weight Prediction on Fetal Ultrasound Video
May 19, 2022



Abstract:Predicting fetal weight at birth is an important aspect of perinatal care, particularly in the context of antenatal management, which includes the planned timing and the mode of delivery. Accurate prediction of weight using prenatal ultrasound is challenging as it requires images of specific fetal body parts during advanced pregnancy which is difficult to capture due to poor quality of images caused by the lack of amniotic fluid. As a consequence, predictions which rely on standard methods often suffer from significant errors. In this paper we propose the Residual Transformer Module which extends a 3D ResNet-based network for analysis of 2D+t spatio-temporal ultrasound video scans. Our end-to-end method, called BabyNet, automatically predicts fetal birth weight based on fetal ultrasound video scans. We evaluate BabyNet using a dedicated clinical set comprising 225 2D fetal ultrasound videos of pregnancies from 75 patients performed one day prior to delivery. Experimental results show that BabyNet outperforms several state-of-the-art methods and estimates the weight at birth with accuracy comparable to human experts. Furthermore, combining estimates provided by human experts with those computed by BabyNet yields the best results, outperforming either of other methods by a significant margin. The source code of BabyNet is available at https://github.com/SanoScience/BabyNet.
FetalNet: Multi-task deep learning framework for fetal ultrasound biometric measurements
Jul 14, 2021
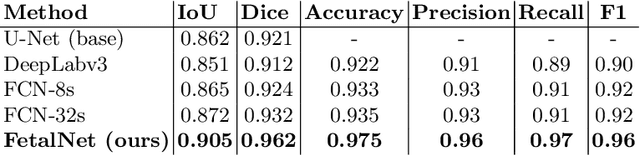
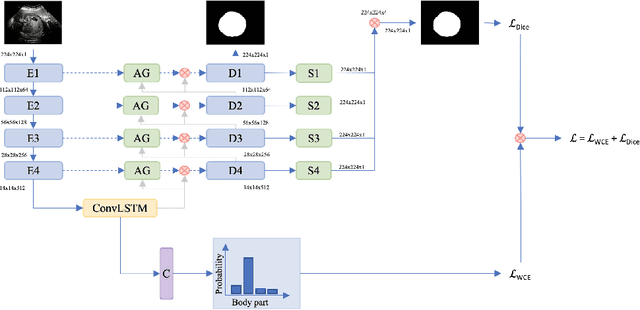
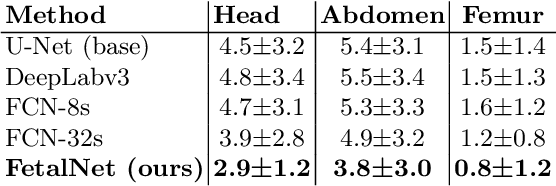
Abstract:In this paper, we propose an end-to-end multi-task neural network called FetalNet with an attention mechanism and stacked module for spatio-temporal fetal ultrasound scan video analysis. Fetal biometric measurement is a standard examination during pregnancy used for the fetus growth monitoring and estimation of gestational age and fetal weight. The main goal in fetal ultrasound scan video analysis is to find proper standard planes to measure the fetal head, abdomen and femur. Due to natural high speckle noise and shadows in ultrasound data, medical expertise and sonographic experience are required to find the appropriate acquisition plane and perform accurate measurements of the fetus. In addition, existing computer-aided methods for fetal US biometric measurement address only one single image frame without considering temporal features. To address these shortcomings, we propose an end-to-end multi-task neural network for spatio-temporal ultrasound scan video analysis to simultaneously localize, classify and measure the fetal body parts. We propose a new encoder-decoder segmentation architecture that incorporates a classification branch. Additionally, we employ an attention mechanism with a stacked module to learn salient maps to suppress irrelevant US regions and efficient scan plane localization. We trained on the fetal ultrasound video comes from routine examinations of 700 different patients. Our method called FetalNet outperforms existing state-of-the-art methods in both classification and segmentation in fetal ultrasound video recordings.
Spontaneous preterm birth prediction using convolutional neural networks
Aug 21, 2020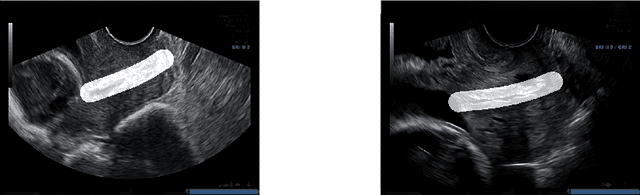
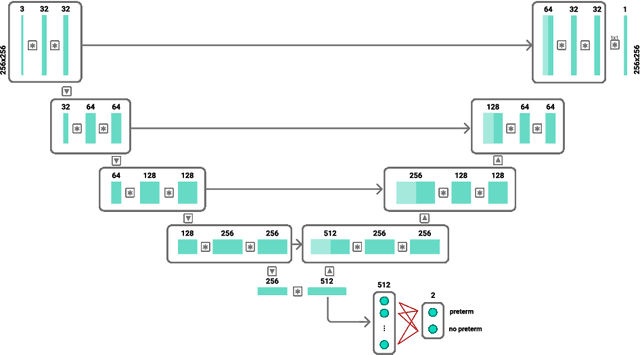
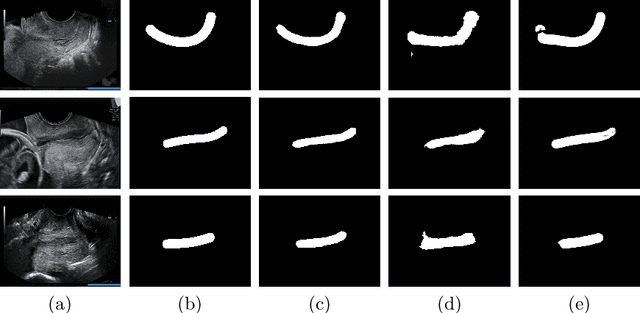
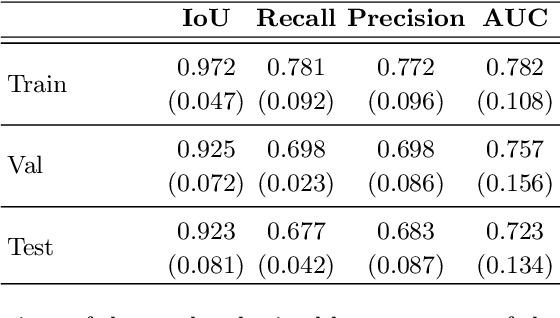
Abstract:An estimated 15 million babies are born too early every year. Approximately 1 million children die each year due to complications of preterm birth (PTB). Many survivors face a lifetime of disability, including learning disabilities and visual and hearing problems. Although manual analysis of ultrasound images (US) is still prevalent, it is prone to errors due to its subjective component and complex variations in the shape and position of organs across patients. In this work, we introduce a conceptually simple convolutional neural network (CNN) trained for segmenting prenatal ultrasound images and classifying task for the purpose of preterm birth detection. Our method efficiently segments different types of cervixes in transvaginal ultrasound images while simultaneously predicting a preterm birth based on extracted image features without human oversight. We employed three popular network models: U-Net, Fully Convolutional Network, and Deeplabv3 for the cervix segmentation task. Based on the conducted results and model efficiency, we decided to extend U-Net by adding a parallel branch for classification task. The proposed model is trained and evaluated on a dataset consisting of 354 2D transvaginal ultrasound images and achieved a segmentation accuracy with a mean Jaccard coefficient index of 0.923 $\pm$ 0.081 and a classification sensitivity of 0.677 $\pm$ 0.042 with a 3.49\% false positive rate. Our method obtained better results in the prediction of preterm birth based on transvaginal ultrasound images compared to state-of-the-art methods.
Estimation of preterm birth markers with U-Net segmentation network
Aug 24, 2019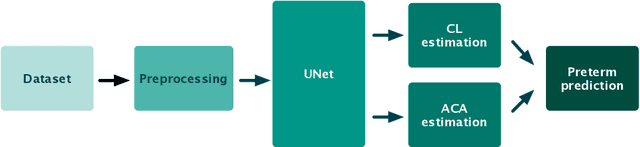
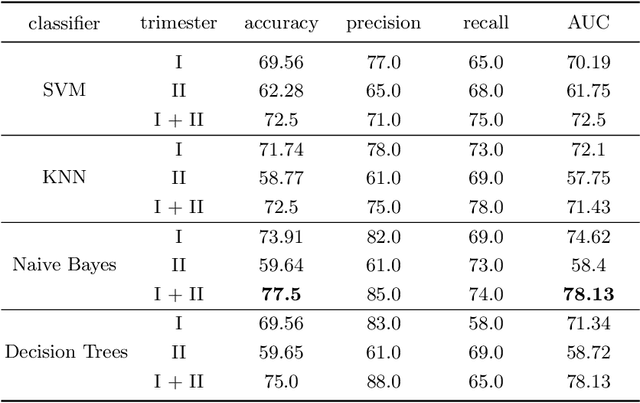
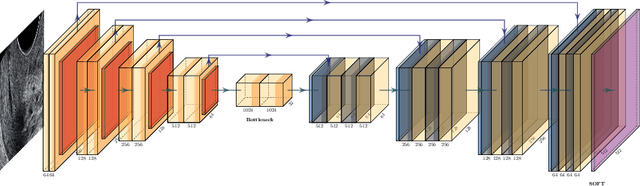
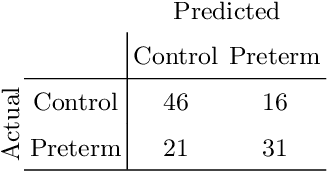
Abstract:Preterm birth is the most common cause of neonatal death. Current diagnostic methods that assess the risk of preterm birth involve the collection of maternal characteristics and transvaginal ultrasound imaging conducted in the first and second trimester of pregnancy. Analysis of the ultrasound data is based on visual inspection of images by gynaecologist, sometimes supported by hand-designed image features such as cervical length. Due to the complexity of this process and its subjective component, approximately 30% of spontaneous preterm deliveries are not correctly predicted. Moreover, 10% of the predicted preterm deliveries are false-positives. In this paper, we address the problem of predicting spontaneous preterm delivery using machine learning. To achieve this goal, we propose to first use a deep neural network architecture for segmenting prenatal ultrasound images and then automatically extract two biophysical ultrasound markers, cervical length (CL) and anterior cervical angle (ACA), from the resulting images. Our method allows to estimate ultrasound markers without human oversight. Furthermore, we show that CL and ACA markers, when combined, allow us to decrease false-negative ratio from 30% to 18%. Finally, contrary to the current approaches to diagnostics methods that rely only on gynaecologist's expertise, our method introduce objectively obtained results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge