Maciej Malawski
Validation of Various Normalization Methods for Brain Tumor Segmentation: Can Federated Learning Overcome This Heterogeneity?
Oct 08, 2025Abstract:Deep learning (DL) has been increasingly applied in medical imaging, however, it requires large amounts of data, which raises many challenges related to data privacy, storage, and transfer. Federated learning (FL) is a training paradigm that overcomes these issues, though its effectiveness may be reduced when dealing with non-independent and identically distributed (non-IID) data. This study simulates non-IID conditions by applying different MRI intensity normalization techniques to separate data subsets, reflecting a common cause of heterogeneity. These subsets are then used for training and testing models for brain tumor segmentation. The findings provide insights into the influence of the MRI intensity normalization methods on segmentation models, both training and inference. Notably, the FL methods demonstrated resilience to inconsistently normalized data across clients, achieving the 3D Dice score of 92%, which is comparable to a centralized model (trained using all data). These results indicate that FL is a solution to effectively train high-performing models without violating data privacy, a crucial concern in medical applications. The code is available at: https://github.com/SanoScience/fl-varying-normalization.
AI Factories: It's time to rethink the Cloud-HPC divide
Sep 16, 2025Abstract:The strategic importance of artificial intelligence is driving a global push toward Sovereign AI initiatives. Nationwide governments are increasingly developing dedicated infrastructures, called AI Factories (AIF), to achieve technological autonomy and secure the resources necessary to sustain robust local digital ecosystems. In Europe, the EuroHPC Joint Undertaking is investing hundreds of millions of euros into several AI Factories, built atop existing high-performance computing (HPC) supercomputers. However, while HPC systems excel in raw performance, they are not inherently designed for usability, accessibility, or serving as public-facing platforms for AI services such as inference or agentic applications. In contrast, AI practitioners are accustomed to cloud-native technologies like Kubernetes and object storage, tools that are often difficult to integrate within traditional HPC environments. This article advocates for a dual-stack approach within supercomputers: integrating both HPC and cloud-native technologies. Our goal is to bridge the divide between HPC and cloud computing by combining high performance and hardware acceleration with ease of use and service-oriented front-ends. This convergence allows each paradigm to amplify the other. To this end, we will study the cloud challenges of HPC (Serverless HPC) and the HPC challenges of cloud technologies (High-performance Cloud).
Federated Learning: A new frontier in the exploration of multi-institutional medical imaging data
Mar 25, 2025Abstract:Artificial intelligence has transformed the perspective of medical imaging, leading to a genuine technological revolution in modern computer-assisted healthcare systems. However, ubiquitously featured deep learning (DL) systems require access to a considerable amount of data, facilitating proper knowledge extraction and generalization. Admission to such extensive resources may be hindered due to the time and effort required to convey ethical agreements, set up and carry the acquisition procedures through, and manage the datasets adequately with a particular emphasis on proper anonymization. One of the pivotal challenges in the DL field is data integration from various sources acquired using different hardware vendors, diverse acquisition protocols, experimental setups, and even inter-operator variabilities. In this paper, we review the federated learning (FL) concept that fosters the integration of large-scale heterogeneous datasets from multiple institutions in training DL models. In contrast to a centralized approach, the decentralized FL procedure promotes training DL models while preserving data privacy at each institution involved. We formulate the FL principle and comprehensively review general and dedicated medical imaging aggregation and learning algorithms, enabling the generation of a globally generalized model. We meticulously go through the challenges in constructing FL-based systems, such as data heterogeneity across the institutions, resilience to potential attacks on data privacy, and the variability in computational and communication resources among the entangled sites that might induce efficiency issues of the entire system. Finally, we explore the up-to-date open frameworks for rapid FL-based algorithm prototyping and shed light on future directions in this intensively growing field.
MICCAI-CDMRI 2023 QuantConn Challenge Findings on Achieving Robust Quantitative Connectivity through Harmonized Preprocessing of Diffusion MRI
Nov 14, 2024



Abstract:White matter alterations are increasingly implicated in neurological diseases and their progression. International-scale studies use diffusion-weighted magnetic resonance imaging (DW-MRI) to qualitatively identify changes in white matter microstructure and connectivity. Yet, quantitative analysis of DW-MRI data is hindered by inconsistencies stemming from varying acquisition protocols. There is a pressing need to harmonize the preprocessing of DW-MRI datasets to ensure the derivation of robust quantitative diffusion metrics across acquisitions. In the MICCAI-CDMRI 2023 QuantConn challenge, participants were provided raw data from the same individuals collected on the same scanner but with two different acquisitions and tasked with preprocessing the DW-MRI to minimize acquisition differences while retaining biological variation. Submissions are evaluated on the reproducibility and comparability of cross-acquisition bundle-wise microstructure measures, bundle shape features, and connectomics. The key innovations of the QuantConn challenge are that (1) we assess bundles and tractography in the context of harmonization for the first time, (2) we assess connectomics in the context of harmonization for the first time, and (3) we have 10x additional subjects over prior harmonization challenge, MUSHAC and 100x over SuperMUDI. We find that bundle surface area, fractional anisotropy, connectome assortativity, betweenness centrality, edge count, modularity, nodal strength, and participation coefficient measures are most biased by acquisition and that machine learning voxel-wise correction, RISH mapping, and NeSH methods effectively reduce these biases. In addition, microstructure measures AD, MD, RD, bundle length, connectome density, efficiency, and path length are least biased by these acquisition differences.
* Accepted for publication at the Journal of Machine Learning for Biomedical Imaging (MELBA) https://melba-journal.org/2024/019
Can gamification reduce the burden of self-reporting in mHealth applications? Feasibility study using machine learning from smartwatch data to estimate cognitive load
Feb 07, 2023Abstract:The effectiveness of digital treatments can be measured by requiring patients to self-report their mental and physical state through mobile applications. However, self-reporting can be overwhelming and may cause patients to disengage from the intervention. In order to address this issue, we conduct a feasibility study to explore the impact of gamification on the cognitive burden of self-reporting. Our approach involves the creation of a system to assess cognitive burden through the analysis of photoplethysmography (PPG) signals obtained from a smartwatch. The system is built by collecting PPG data during both cognitively demanding tasks and periods of rest. The obtained data is utilized to train a machine learning model to detect cognitive load (CL). Subsequently, we create two versions of health surveys: a gamified version and a traditional version. Our aim is to estimate the cognitive load experienced by participants while completing these surveys using their mobile devices. We find that CL detector performance can be enhanced via pre-training on stress detection tasks and requires capturing of a minimum 30 seconds of PPG signal to work adequately. For 10 out of 13 participants, a personalized cognitive load detector can achieve an F1 score above 0.7. We find no difference between the gamified and non-gamified mobile surveys in terms of time spent in the state of high cognitive load but participants prefer the gamified version. The average time spent on each question is 5.5 for gamified survey vs 6 seconds for the non-gamified version.
CXR-FL: Deep Learning-based Chest X-ray Image Analysis Using Federated Learning
Apr 11, 2022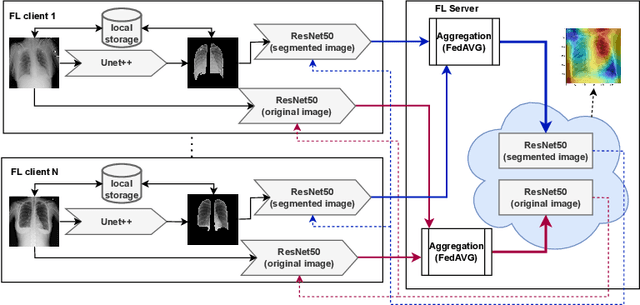
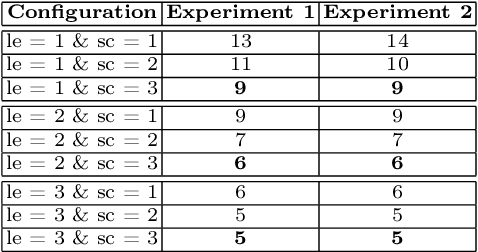
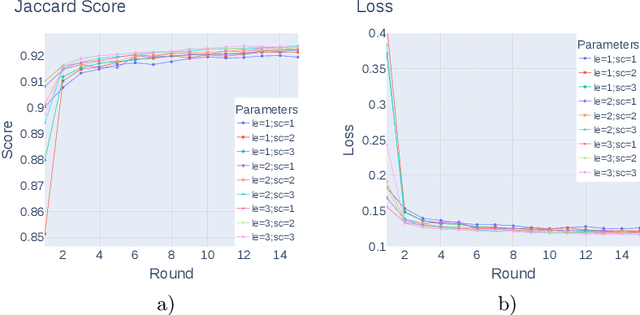
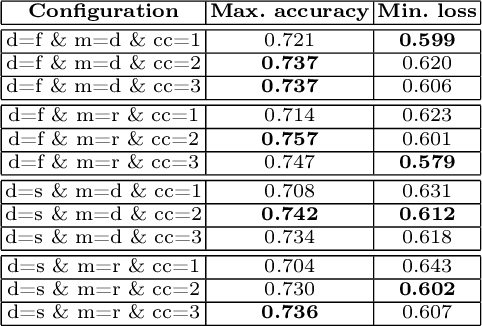
Abstract:Federated learning enables building a shared model from multicentre data while storing the training data locally for privacy. In this paper, we present an evaluation (called CXR-FL) of deep learning-based models for chest X-ray image analysis using the federated learning method. We examine the impact of federated learning parameters on the performance of central models. Additionally, we show that classification models perform worse if trained on a region of interest reduced to segmentation of the lung compared to the full image. However, focusing training of the classification model on the lung area may result in improved pathology interpretability during inference. We also find that federated learning helps maintain model generalizability. The pre-trained weights and code are publicly available at (https://github.com/SanoScience/CXR-FL).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge