Maciej Pajak
Paying Per-label Attention for Multi-label Extraction from Radiology Reports
Aug 07, 2020Abstract:Training medical image analysis models requires large amounts of expertly annotated data which is time-consuming and expensive to obtain. Images are often accompanied by free-text radiology reports which are a rich source of information. In this paper, we tackle the automated extraction of structured labels from head CT reports for imaging of suspected stroke patients, using deep learning. Firstly, we propose a set of 31 labels which correspond to radiographic findings (e.g. hyperdensity) and clinical impressions (e.g. haemorrhage) related to neurological abnormalities. Secondly, inspired by previous work, we extend existing state-of-the-art neural network models with a label-dependent attention mechanism. Using this mechanism and simple synthetic data augmentation, we are able to robustly extract many labels with a single model, classified according to the radiologist's reporting (positive, uncertain, negative). This approach can be used in further research to effectively extract many labels from medical text.
Teacher-Student chain for efficient semi-supervised histology image classification
Mar 20, 2020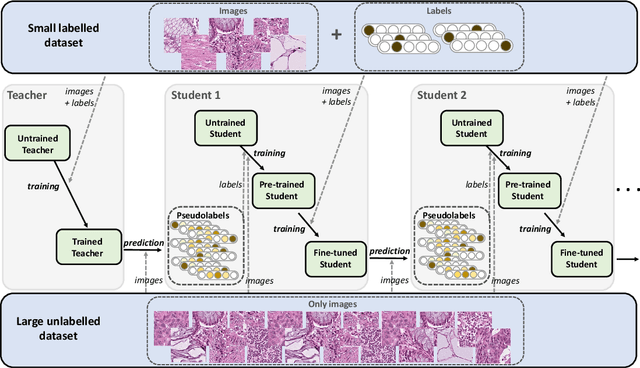

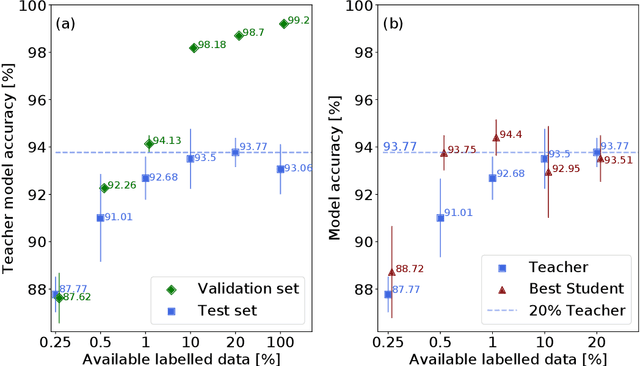
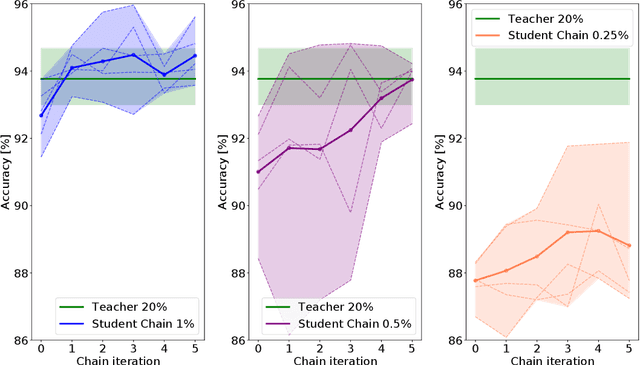
Abstract:Deep learning shows great potential for the domain of digital pathology. An automated digital pathology system could serve as a second reader, perform initial triage in large screening studies, or assist in reporting. However, it is expensive to exhaustively annotate large histology image databases, since medical specialists are a scarce resource. In this paper, we apply the semi-supervised teacher-student knowledge distillation technique proposed by Yalniz et al. (2019) to the task of quantifying prognostic features in colorectal cancer. We obtain accuracy improvements through extending this approach to a chain of students, where each student's predictions are used to train the next student i.e. the student becomes the teacher. Using the chain approach, and only 0.5% labelled data (the remaining 99.5% in the unlabelled pool), we match the accuracy of training on 100% labelled data. At lower percentages of labelled data, similar gains in accuracy are seen, allowing some recovery of accuracy even from a poor initial choice of labelled training set. In conclusion, this approach shows promise for reducing the annotation burden, thus increasing the affordability of automated digital pathology systems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge