Ziyuan Zhao
Addressing Personalized Bias for Unbiased Learning to Rank
Aug 28, 2025Abstract:Unbiased learning to rank (ULTR), which aims to learn unbiased ranking models from biased user behavior logs, plays an important role in Web search. Previous research on ULTR has studied a variety of biases in users' clicks, such as position bias, presentation bias, and outlier bias. However, existing work often assumes that the behavior logs are collected from an ``average'' user, neglecting the differences between different users in their search and browsing behaviors. In this paper, we introduce personalized factors into the ULTR framework, which we term the user-aware ULTR problem. Through a formal causal analysis of this problem, we demonstrate that existing user-oblivious methods are biased when different users have different preferences over queries and personalized propensities of examining documents. To address such a personalized bias, we propose a novel user-aware inverse-propensity-score estimator for learning-to-rank objectives. Specifically, our approach models the distribution of user browsing behaviors for each query and aggregates user-weighted examination probabilities to determine propensities. We theoretically prove that the user-aware estimator is unbiased under some mild assumptions and shows lower variance compared to the straightforward way of calculating a user-dependent propensity for each impression. Finally, we empirically verify the effectiveness of our user-aware estimator by conducting extensive experiments on two semi-synthetic datasets and a real-world dataset.
crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023
Jun 13, 2025Abstract:The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
Energy-efficient Hybrid Model Predictive Trajectory Planning for Autonomous Electric Vehicles
Nov 09, 2024Abstract:To tackle the twin challenges of limited battery life and lengthy charging durations in electric vehicles (EVs), this paper introduces an Energy-efficient Hybrid Model Predictive Planner (EHMPP), which employs an energy-saving optimization strategy. EHMPP focuses on refining the design of the motion planner to be seamlessly integrated with the existing automatic driving algorithms, without additional hardware. It has been validated through simulation experiments on the Prescan, CarSim, and Matlab platforms, demonstrating that it can increase passive recovery energy by 11.74\% and effectively track motor speed and acceleration at optimal power. To sum up, EHMPP not only aids in trajectory planning but also significantly boosts energy efficiency in autonomous EVs.
Graph Neural Networks for Protein-Protein Interactions -- A Short Survey
Apr 16, 2024Abstract:Protein-protein interactions (PPIs) play key roles in a broad range of biological processes. Numerous strategies have been proposed for predicting PPIs, and among them, graph-based methods have demonstrated promising outcomes owing to the inherent graph structure of PPI networks. This paper reviews various graph-based methodologies, and discusses their applications in PPI prediction. We classify these approaches into two primary groups based on their model structures. The first category employs Graph Neural Networks (GNN) or Graph Convolutional Networks (GCN), while the second category utilizes Graph Attention Networks (GAT), Graph Auto-Encoders and Graph-BERT. We highlight the distinctive methodologies of each approach in managing the graph-structured data inherent in PPI networks and anticipate future research directions in this domain.
SemiGNN-PPI: Self-Ensembling Multi-Graph Neural Network for Efficient and Generalizable Protein-Protein Interaction Prediction
May 15, 2023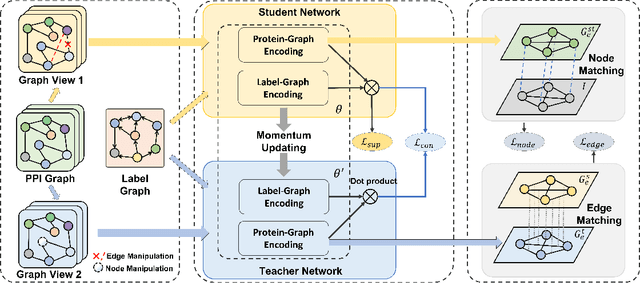
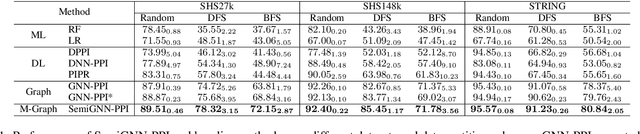
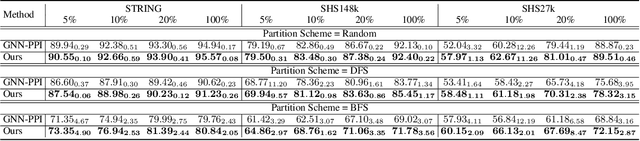
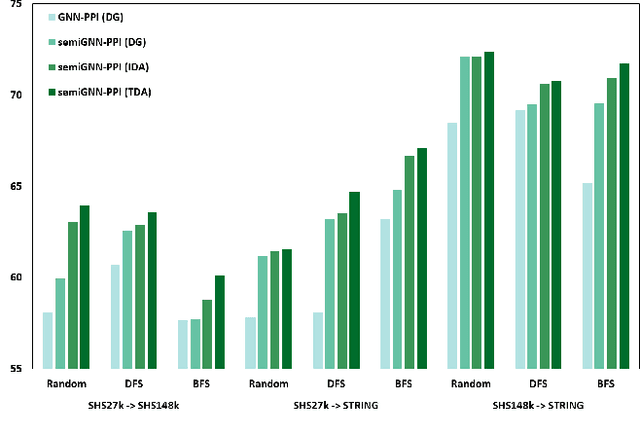
Abstract:Protein-protein interactions (PPIs) are crucial in various biological processes and their study has significant implications for drug development and disease diagnosis. Existing deep learning methods suffer from significant performance degradation under complex real-world scenarios due to various factors, e.g., label scarcity and domain shift. In this paper, we propose a self-ensembling multigraph neural network (SemiGNN-PPI) that can effectively predict PPIs while being both efficient and generalizable. In SemiGNN-PPI, we not only model the protein correlations but explore the label dependencies by constructing and processing multiple graphs from the perspectives of both features and labels in the graph learning process. We further marry GNN with Mean Teacher to effectively leverage unlabeled graph-structured PPI data for self-ensemble graph learning. We also design multiple graph consistency constraints to align the student and teacher graphs in the feature embedding space, enabling the student model to better learn from the teacher model by incorporating more relationships. Extensive experiments on PPI datasets of different scales with different evaluation settings demonstrate that SemiGNN-PPI outperforms state-of-the-art PPI prediction methods, particularly in challenging scenarios such as training with limited annotations and testing on unseen data.
Meta-hallucinator: Towards Few-Shot Cross-Modality Cardiac Image Segmentation
May 11, 2023Abstract:Domain shift and label scarcity heavily limit deep learning applications to various medical image analysis tasks. Unsupervised domain adaptation (UDA) techniques have recently achieved promising cross-modality medical image segmentation by transferring knowledge from a label-rich source domain to an unlabeled target domain. However, it is also difficult to collect annotations from the source domain in many clinical applications, rendering most prior works suboptimal with the label-scarce source domain, particularly for few-shot scenarios, where only a few source labels are accessible. To achieve efficient few-shot cross-modality segmentation, we propose a novel transformation-consistent meta-hallucination framework, meta-hallucinator, with the goal of learning to diversify data distributions and generate useful examples for enhancing cross-modality performance. In our framework, hallucination and segmentation models are jointly trained with the gradient-based meta-learning strategy to synthesize examples that lead to good segmentation performance on the target domain. To further facilitate data hallucination and cross-domain knowledge transfer, we develop a self-ensembling model with a hallucination-consistent property. Our meta-hallucinator can seamlessly collaborate with the meta-segmenter for learning to hallucinate with mutual benefits from a combined view of meta-learning and self-ensembling learning. Extensive studies on MM-WHS 2017 dataset for cross-modality cardiac segmentation demonstrate that our method performs favorably against various approaches by a lot in the few-shot UDA scenario.
* Accepted by MICCAI 2022 (top 13% paper; early accept)
MS-MT: Multi-Scale Mean Teacher with Contrastive Unpaired Translation for Cross-Modality Vestibular Schwannoma and Cochlea Segmentation
Mar 28, 2023Abstract:Domain shift has been a long-standing issue for medical image segmentation. Recently, unsupervised domain adaptation (UDA) methods have achieved promising cross-modality segmentation performance by distilling knowledge from a label-rich source domain to a target domain without labels. In this work, we propose a multi-scale self-ensembling based UDA framework for automatic segmentation of two key brain structures i.e., Vestibular Schwannoma (VS) and Cochlea on high-resolution T2 images. First, a segmentation-enhanced contrastive unpaired image translation module is designed for image-level domain adaptation from source T1 to target T2. Next, multi-scale deep supervision and consistency regularization are introduced to a mean teacher network for self-ensemble learning to further close the domain gap. Furthermore, self-training and intensity augmentation techniques are utilized to mitigate label scarcity and boost cross-modality segmentation performance. Our method demonstrates promising segmentation performance with a mean Dice score of 83.8% and 81.4% and an average asymmetric surface distance (ASSD) of 0.55 mm and 0.26 mm for the VS and Cochlea, respectively in the validation phase of the crossMoDA 2022 challenge.
Multimodal Continuous Emotion Recognition: A Technical Report for ABAW5
Mar 18, 2023


Abstract:We used two multimodal models for continuous valence-arousal recognition using visual, audio, and linguistic information. The first model is the same as we used in ABAW2 and ABAW3, which employs the leader-follower attention. The second model has the same architecture for spatial and temporal encoding. As for the fusion block, it employs a compact and straightforward channel attention, borrowed from the End2You toolkit. Unlike our previous attempts that use Vggish feature directly as the audio feature, this time we feed the pre-trained VGG model using logmel-spectrogram and finetune it during the training. To make full use of the data and alleviate over-fitting, cross-validation is carried out. The fold with the highest concordance correlation coefficient is selected for submission. The code is to be available at https://github.com/sucv/ABAW5.
MetaGrad: Adaptive Gradient Quantization with Hypernetworks
Mar 04, 2023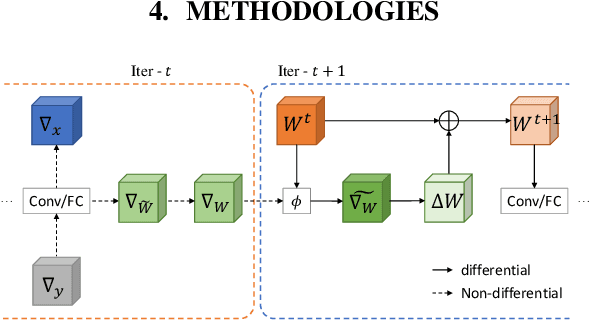

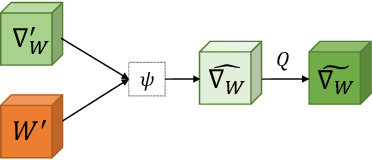

Abstract:A popular track of network compression approach is Quantization aware Training (QAT), which accelerates the forward pass during the neural network training and inference. However, not much prior efforts have been made to quantize and accelerate the backward pass during training, even though that contributes around half of the training time. This can be partly attributed to the fact that errors of low-precision gradients during backward cannot be amortized by the training objective as in the QAT setting. In this work, we propose to solve this problem by incorporating the gradients into the computation graph of the next training iteration via a hypernetwork. Various experiments on CIFAR-10 dataset with different CNN network architectures demonstrate that our hypernetwork-based approach can effectively reduce the negative effect of gradient quantization noise and successfully quantizes the gradients to INT4 with only 0.64 accuracy drop for VGG-16 on CIFAR-10.
DA-CIL: Towards Domain Adaptive Class-Incremental 3D Object Detection
Dec 05, 2022



Abstract:Deep learning has achieved notable success in 3D object detection with the advent of large-scale point cloud datasets. However, severe performance degradation in the past trained classes, i.e., catastrophic forgetting, still remains a critical issue for real-world deployment when the number of classes is unknown or may vary. Moreover, existing 3D class-incremental detection methods are developed for the single-domain scenario, which fail when encountering domain shift caused by different datasets, varying environments, etc. In this paper, we identify the unexplored yet valuable scenario, i.e., class-incremental learning under domain shift, and propose a novel 3D domain adaptive class-incremental object detection framework, DA-CIL, in which we design a novel dual-domain copy-paste augmentation method to construct multiple augmented domains for diversifying training distributions, thereby facilitating gradual domain adaptation. Then, multi-level consistency is explored to facilitate dual-teacher knowledge distillation from different domains for domain adaptive class-incremental learning. Extensive experiments on various datasets demonstrate the effectiveness of the proposed method over baselines in the domain adaptive class-incremental learning scenario.
* Accepted by the 33rd British Machine Vision Conference (BMVC 2022)
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge