Zeng Zeng
Institute for Infocomm Research
Utilizing the Mean Teacher with Supcontrast Loss for Wafer Pattern Recognition
Nov 27, 2024Abstract:The patterns on wafer maps play a crucial role in helping engineers identify the causes of production issues during semiconductor manufacturing. In order to reduce costs and improve accuracy, automation technology is essential, and recent developments in deep learning have led to impressive results in wafer map pattern recognition. In this context, inspired by the effectiveness of semi-supervised learning and contrastive learning methods, we introduce an innovative approach that integrates the Mean Teacher framework with the supervised contrastive learning loss for enhanced wafer map pattern recognition. Our methodology not only addresses the nuances of wafer patterns but also tackles challenges arising from limited labeled data. To further refine the process, we address data imbalance in the wafer dataset by employing SMOTE and under-sampling techniques. We conduct a comprehensive analysis of our proposed method and demonstrate its effectiveness through experiments using real-world dataset WM811K obtained from semiconductor manufacturers. Compared to the baseline method, our method has achieved 5.46%, 6.68%, 5.42%, and 4.53% improvements in Accuracy, Precision, Recall, and F1 score, respectively.
Graph Neural Networks for Protein-Protein Interactions -- A Short Survey
Apr 16, 2024Abstract:Protein-protein interactions (PPIs) play key roles in a broad range of biological processes. Numerous strategies have been proposed for predicting PPIs, and among them, graph-based methods have demonstrated promising outcomes owing to the inherent graph structure of PPI networks. This paper reviews various graph-based methodologies, and discusses their applications in PPI prediction. We classify these approaches into two primary groups based on their model structures. The first category employs Graph Neural Networks (GNN) or Graph Convolutional Networks (GCN), while the second category utilizes Graph Attention Networks (GAT), Graph Auto-Encoders and Graph-BERT. We highlight the distinctive methodologies of each approach in managing the graph-structured data inherent in PPI networks and anticipate future research directions in this domain.
A Deeply Supervised Semantic Segmentation Method Based on GAN
Oct 06, 2023Abstract:In recent years, the field of intelligent transportation has witnessed rapid advancements, driven by the increasing demand for automation and efficiency in transportation systems. Traffic safety, one of the tasks integral to intelligent transport systems, requires accurately identifying and locating various road elements, such as road cracks, lanes, and traffic signs. Semantic segmentation plays a pivotal role in achieving this task, as it enables the partition of images into meaningful regions with accurate boundaries. In this study, we propose an improved semantic segmentation model that combines the strengths of adversarial learning with state-of-the-art semantic segmentation techniques. The proposed model integrates a generative adversarial network (GAN) framework into the traditional semantic segmentation model, enhancing the model's performance in capturing complex and subtle features in transportation images. The effectiveness of our approach is demonstrated by a significant boost in performance on the road crack dataset compared to the existing methods, \textit{i.e.,} SEGAN. This improvement can be attributed to the synergistic effect of adversarial learning and semantic segmentation, which leads to a more refined and accurate representation of road structures and conditions. The enhanced model not only contributes to better detection of road cracks but also to a wide range of applications in intelligent transportation, such as traffic sign recognition, vehicle detection, and lane segmentation.
SemiGNN-PPI: Self-Ensembling Multi-Graph Neural Network for Efficient and Generalizable Protein-Protein Interaction Prediction
May 15, 2023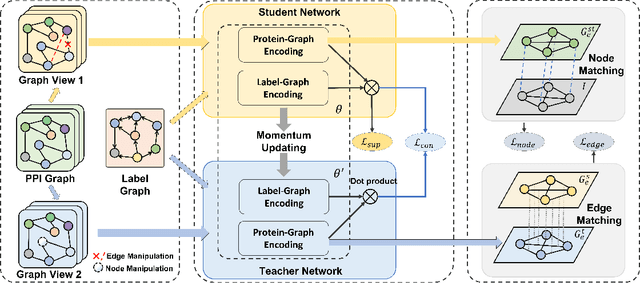
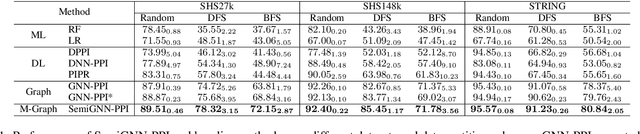
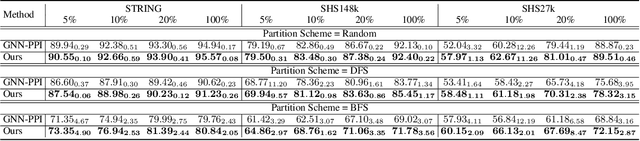
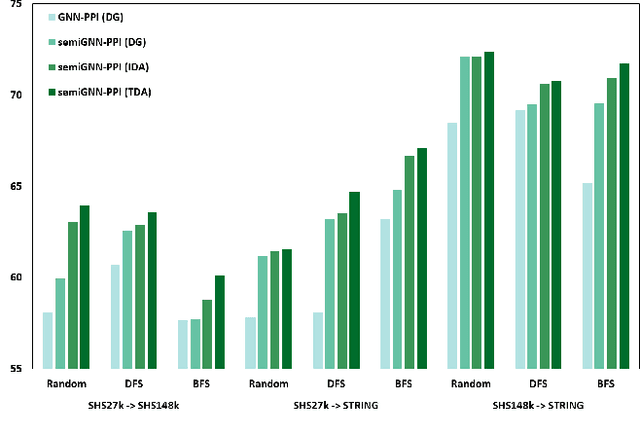
Abstract:Protein-protein interactions (PPIs) are crucial in various biological processes and their study has significant implications for drug development and disease diagnosis. Existing deep learning methods suffer from significant performance degradation under complex real-world scenarios due to various factors, e.g., label scarcity and domain shift. In this paper, we propose a self-ensembling multigraph neural network (SemiGNN-PPI) that can effectively predict PPIs while being both efficient and generalizable. In SemiGNN-PPI, we not only model the protein correlations but explore the label dependencies by constructing and processing multiple graphs from the perspectives of both features and labels in the graph learning process. We further marry GNN with Mean Teacher to effectively leverage unlabeled graph-structured PPI data for self-ensemble graph learning. We also design multiple graph consistency constraints to align the student and teacher graphs in the feature embedding space, enabling the student model to better learn from the teacher model by incorporating more relationships. Extensive experiments on PPI datasets of different scales with different evaluation settings demonstrate that SemiGNN-PPI outperforms state-of-the-art PPI prediction methods, particularly in challenging scenarios such as training with limited annotations and testing on unseen data.
Meta-hallucinator: Towards Few-Shot Cross-Modality Cardiac Image Segmentation
May 11, 2023Abstract:Domain shift and label scarcity heavily limit deep learning applications to various medical image analysis tasks. Unsupervised domain adaptation (UDA) techniques have recently achieved promising cross-modality medical image segmentation by transferring knowledge from a label-rich source domain to an unlabeled target domain. However, it is also difficult to collect annotations from the source domain in many clinical applications, rendering most prior works suboptimal with the label-scarce source domain, particularly for few-shot scenarios, where only a few source labels are accessible. To achieve efficient few-shot cross-modality segmentation, we propose a novel transformation-consistent meta-hallucination framework, meta-hallucinator, with the goal of learning to diversify data distributions and generate useful examples for enhancing cross-modality performance. In our framework, hallucination and segmentation models are jointly trained with the gradient-based meta-learning strategy to synthesize examples that lead to good segmentation performance on the target domain. To further facilitate data hallucination and cross-domain knowledge transfer, we develop a self-ensembling model with a hallucination-consistent property. Our meta-hallucinator can seamlessly collaborate with the meta-segmenter for learning to hallucinate with mutual benefits from a combined view of meta-learning and self-ensembling learning. Extensive studies on MM-WHS 2017 dataset for cross-modality cardiac segmentation demonstrate that our method performs favorably against various approaches by a lot in the few-shot UDA scenario.
* Accepted by MICCAI 2022 (top 13% paper; early accept)
LE-UDA: Label-efficient unsupervised domain adaptation for medical image segmentation
Dec 05, 2022Abstract:While deep learning methods hitherto have achieved considerable success in medical image segmentation, they are still hampered by two limitations: (i) reliance on large-scale well-labeled datasets, which are difficult to curate due to the expert-driven and time-consuming nature of pixel-level annotations in clinical practices, and (ii) failure to generalize from one domain to another, especially when the target domain is a different modality with severe domain shifts. Recent unsupervised domain adaptation~(UDA) techniques leverage abundant labeled source data together with unlabeled target data to reduce the domain gap, but these methods degrade significantly with limited source annotations. In this study, we address this underexplored UDA problem, investigating a challenging but valuable realistic scenario, where the source domain not only exhibits domain shift~w.r.t. the target domain but also suffers from label scarcity. In this regard, we propose a novel and generic framework called ``Label-Efficient Unsupervised Domain Adaptation"~(LE-UDA). In LE-UDA, we construct self-ensembling consistency for knowledge transfer between both domains, as well as a self-ensembling adversarial learning module to achieve better feature alignment for UDA. To assess the effectiveness of our method, we conduct extensive experiments on two different tasks for cross-modality segmentation between MRI and CT images. Experimental results demonstrate that the proposed LE-UDA can efficiently leverage limited source labels to improve cross-domain segmentation performance, outperforming state-of-the-art UDA approaches in the literature. Code is available at: https://github.com/jacobzhaoziyuan/LE-UDA.
ACT-Net: Asymmetric Co-Teacher Network for Semi-supervised Memory-efficient Medical Image Segmentation
Jul 05, 2022



Abstract:While deep models have shown promising performance in medical image segmentation, they heavily rely on a large amount of well-annotated data, which is difficult to access, especially in clinical practice. On the other hand, high-accuracy deep models usually come in large model sizes, limiting their employment in real scenarios. In this work, we propose a novel asymmetric co-teacher framework, ACT-Net, to alleviate the burden on both expensive annotations and computational costs for semi-supervised knowledge distillation. We advance teacher-student learning with a co-teacher network to facilitate asymmetric knowledge distillation from large models to small ones by alternating student and teacher roles, obtaining tiny but accurate models for clinical employment. To verify the effectiveness of our ACT-Net, we employ the ACDC dataset for cardiac substructure segmentation in our experiments. Extensive experimental results demonstrate that ACT-Net outperforms other knowledge distillation methods and achieves lossless segmentation performance with 250x fewer parameters.
MMGL: Multi-Scale Multi-View Global-Local Contrastive learning for Semi-supervised Cardiac Image Segmentation
Jul 05, 2022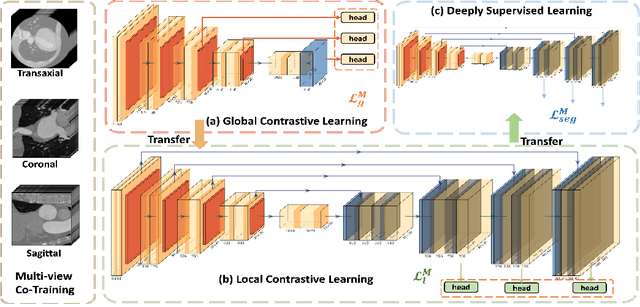

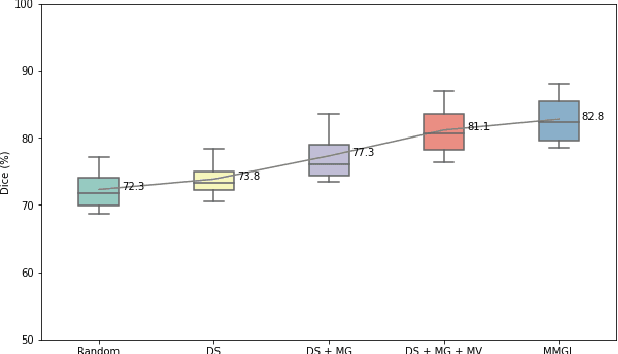
Abstract:With large-scale well-labeled datasets, deep learning has shown significant success in medical image segmentation. However, it is challenging to acquire abundant annotations in clinical practice due to extensive expertise requirements and costly labeling efforts. Recently, contrastive learning has shown a strong capacity for visual representation learning on unlabeled data, achieving impressive performance rivaling supervised learning in many domains. In this work, we propose a novel multi-scale multi-view global-local contrastive learning (MMGL) framework to thoroughly explore global and local features from different scales and views for robust contrastive learning performance, thereby improving segmentation performance with limited annotations. Extensive experiments on the MM-WHS dataset demonstrate the effectiveness of MMGL framework on semi-supervised cardiac image segmentation, outperforming the state-of-the-art contrastive learning methods by a large margin.
Residual Channel Attention Network for Brain Glioma Segmentation
May 22, 2022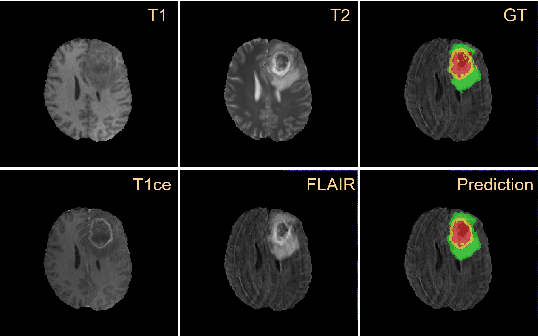
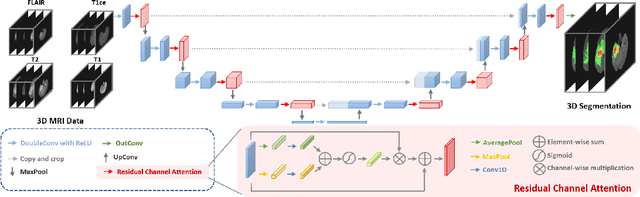
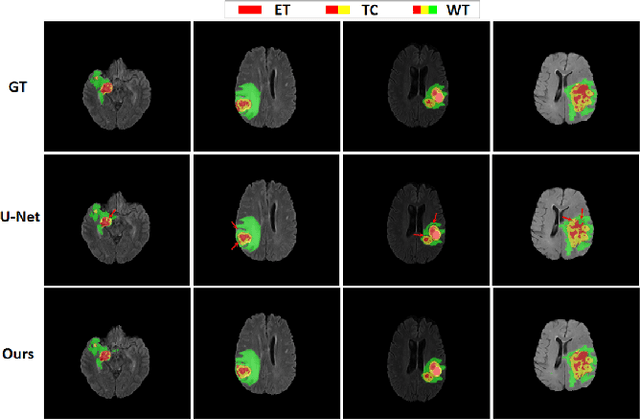
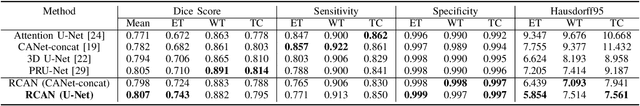
Abstract:A glioma is a malignant brain tumor that seriously affects cognitive functions and lowers patients' life quality. Segmentation of brain glioma is challenging because of interclass ambiguities in tumor regions. Recently, deep learning approaches have achieved outstanding performance in the automatic segmentation of brain glioma. However, existing algorithms fail to exploit channel-wise feature interdependence to select semantic attributes for glioma segmentation. In this study, we implement a novel deep neural network that integrates residual channel attention modules to calibrate intermediate features for glioma segmentation. The proposed channel attention mechanism adaptively weights feature channel-wise to optimize the latent representation of gliomas. We evaluate our method on the established dataset BraTS2017. Experimental results indicate the superiority of our method.
Deep Feature Fusion via Graph Convolutional Network for Intracranial Artery Labeling
May 22, 2022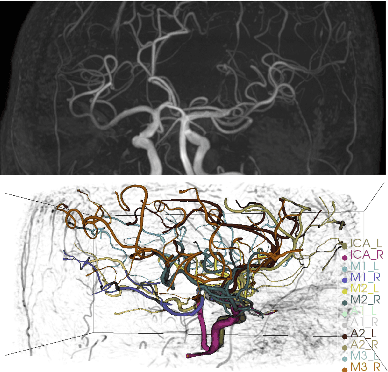
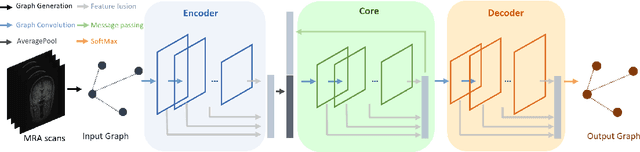

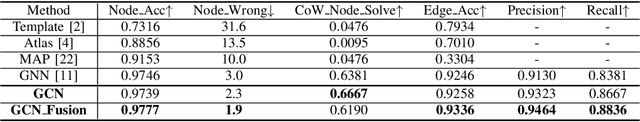
Abstract:Intracranial arteries are critical blood vessels that supply the brain with oxygenated blood. Intracranial artery labels provide valuable guidance and navigation to numerous clinical applications and disease diagnoses. Various machine learning algorithms have been carried out for automation in the anatomical labeling of cerebral arteries. However, the task remains challenging because of the high complexity and variations of intracranial arteries. This study investigates a novel graph convolutional neural network with deep feature fusion for cerebral artery labeling. We introduce stacked graph convolutions in an encoder-core-decoder architecture, extracting high-level representations from graph nodes and their neighbors. Furthermore, we efficiently aggregate intermediate features from different hierarchies to enhance the proposed model's representation capability and labeling performance. We perform extensive experiments on public datasets, in which the results prove the superiority of our approach over baselines by a clear margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge