Yunsheng Bai
AssertionForge: Enhancing Formal Verification Assertion Generation with Structured Representation of Specifications and RTL
Mar 24, 2025



Abstract:Generating SystemVerilog Assertions (SVAs) from natural language specifications remains a major challenge in formal verification (FV) due to the inherent ambiguity and incompleteness of specifications. Existing LLM-based approaches, such as AssertLLM, focus on extracting information solely from specification documents, often failing to capture essential internal signal interactions and design details present in the RTL code, leading to incomplete or incorrect assertions. We propose a novel approach that constructs a Knowledge Graph (KG) from both specifications and RTL, using a hardware-specific schema with domain-specific entity and relation types. We create an initial KG from the specification and then systematically fuse it with information extracted from the RTL code, resulting in a unified, comprehensive KG. This combined representation enables a more thorough understanding of the design and allows for a multi-resolution context synthesis process which is designed to extract diverse verification contexts from the KG. Experiments on four designs demonstrate that our method significantly enhances SVA quality over prior methods. This structured representation not only improves FV but also paves the way for future research in tasks like code generation and design understanding.
ChipAlign: Instruction Alignment in Large Language Models for Chip Design via Geodesic Interpolation
Dec 15, 2024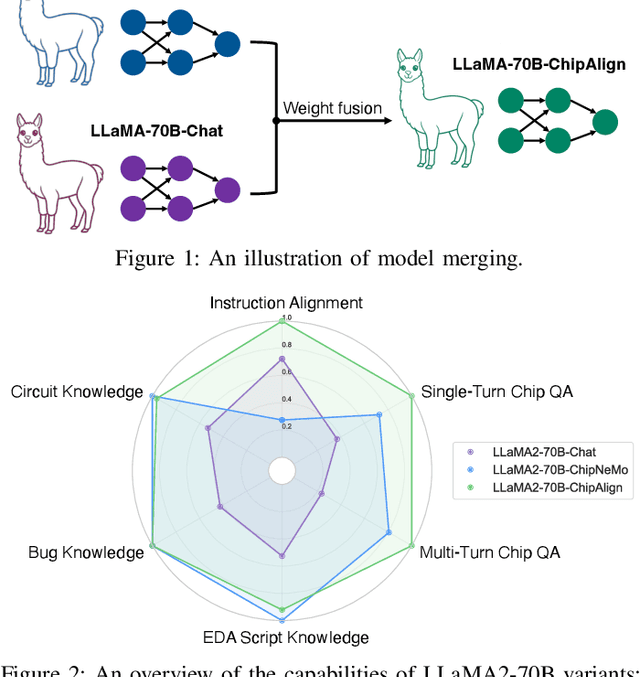
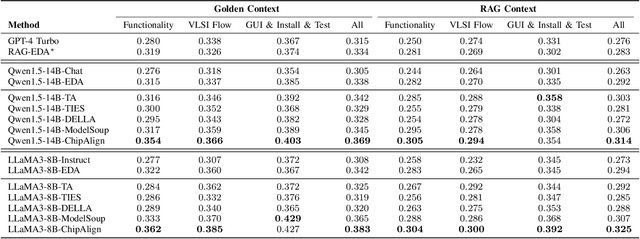

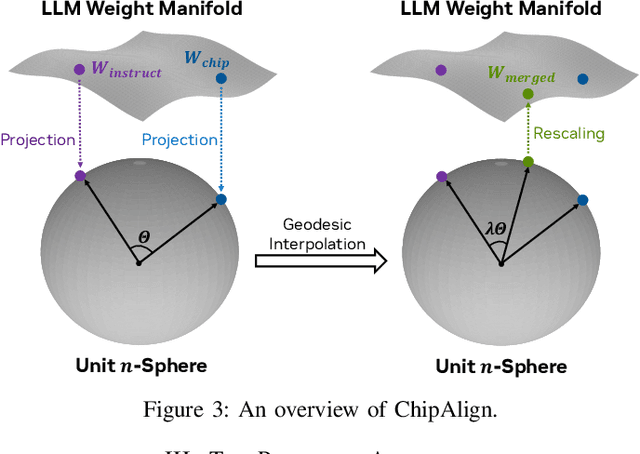
Abstract:Recent advancements in large language models (LLMs) have expanded their application across various domains, including chip design, where domain-adapted chip models like ChipNeMo have emerged. However, these models often struggle with instruction alignment, a crucial capability for LLMs that involves following explicit human directives. This limitation impedes the practical application of chip LLMs, including serving as assistant chatbots for hardware design engineers. In this work, we introduce ChipAlign, a novel approach that utilizes a training-free model merging strategy, combining the strengths of a general instruction-aligned LLM with a chip-specific LLM. By considering the underlying manifold in the weight space, ChipAlign employs geodesic interpolation to effectively fuse the weights of input LLMs, producing a merged model that inherits strong instruction alignment and chip expertise from the respective instruction and chip LLMs. Our results demonstrate that ChipAlign significantly enhances instruction-following capabilities of existing chip LLMs, achieving up to a 26.6% improvement on the IFEval benchmark, while maintaining comparable expertise in the chip domain. This improvement in instruction alignment also translates to notable gains in instruction-involved QA tasks, delivering performance enhancements of 3.9% on the OpenROAD QA benchmark and 8.25% on production-level chip QA benchmarks, surpassing state-of-the-art baselines.
Does Few-Shot Learning Help LLM Performance in Code Synthesis?
Dec 03, 2024



Abstract:Large language models (LLMs) have made significant strides at code generation through improved model design, training, and chain-of-thought. However, prompt-level optimizations remain an important yet under-explored aspect of LLMs for coding. This work focuses on the few-shot examples present in most code generation prompts, offering a systematic study on whether few-shot examples improve LLM's coding capabilities, which few-shot examples have the largest impact, and how to select impactful examples. Our work offers 2 approaches for selecting few-shot examples, a model-free method, CODEEXEMPLAR-FREE, and a model-based method, CODEEXEMPLAR-BASED. The 2 methods offer a trade-off between improved performance and reliance on training data and interpretability. Both methods significantly improve CodeLlama's coding ability across the popular HumanEval+ coding benchmark. In summary, our work provides valuable insights into how to pick few-shot examples in code generation prompts to improve LLM code generation capabilities.
Cross-Modality Program Representation Learning for Electronic Design Automation with High-Level Synthesis
Jun 13, 2024Abstract:In recent years, domain-specific accelerators (DSAs) have gained popularity for applications such as deep learning and autonomous driving. To facilitate DSA designs, programmers use high-level synthesis (HLS) to compile a high-level description written in C/C++ into a design with low-level hardware description languages that eventually synthesize DSAs on circuits. However, creating a high-quality HLS design still demands significant domain knowledge, particularly in microarchitecture decisions expressed as \textit{pragmas}. Thus, it is desirable to automate such decisions with the help of machine learning for predicting the quality of HLS designs, requiring a deeper understanding of the program that consists of original code and pragmas. Naturally, these programs can be considered as sequence data. In addition, these programs can be compiled and converted into a control data flow graph (CDFG). But existing works either fail to leverage both modalities or combine the two in shallow or coarse ways. We propose ProgSG, a model that allows interaction between the source code sequence modality and the graph modality in a deep and fine-grained way. To alleviate the scarcity of labeled designs, a pre-training method is proposed based on a suite of compiler's data flow analysis tasks. Experimental results show that ProgSG reduces the RMSE of design performance predictions by up to $22\%$, and identifies designs with an average of $1.10\times$ and $1.26\times$ (up to $8.17\times$ and $13.31\times$) performance improvement in design space exploration (DSE) task compared to HARP and AutoDSE, respectively.
ProgSG: Cross-Modality Representation Learning for Programs in Electronic Design Automation
May 18, 2023



Abstract:Recent years have witnessed the growing popularity of domain-specific accelerators (DSAs), such as Google's TPUs, for accelerating various applications such as deep learning, search, autonomous driving, etc. To facilitate DSA designs, high-level synthesis (HLS) is used, which allows a developer to compile a high-level description in the form of software code in C and C++ into a design in low-level hardware description languages (such as VHDL or Verilog) and eventually synthesized into a DSA on an ASIC (application-specific integrated circuit) or FPGA (field-programmable gate arrays). However, existing HLS tools still require microarchitecture decisions, expressed in terms of pragmas (such as directives for parallelization and pipelining). To enable more people to design DSAs, it is desirable to automate such decisions with the help of deep learning for predicting the quality of HLS designs. This requires us a deeper understanding of the program, which is a combination of original code and pragmas. Naturally, these programs can be considered as sequence data, for which large language models (LLM) can help. In addition, these programs can be compiled and converted into a control data flow graph (CDFG), and the compiler also provides fine-grained alignment between the code tokens and the CDFG nodes. However, existing works either fail to leverage both modalities or combine the two in shallow or coarse ways. We propose ProgSG allowing the source code sequence modality and the graph modalities to interact with each other in a deep and fine-grained way. To alleviate the scarcity of labeled designs, a pre-training method is proposed based on a suite of compiler's data flow analysis tasks. Experimental results on two benchmark datasets show the superiority of ProgSG over baseline methods that either only consider one modality or combine the two without utilizing the alignment information.
Code Recommendation for Open Source Software Developers
Oct 20, 2022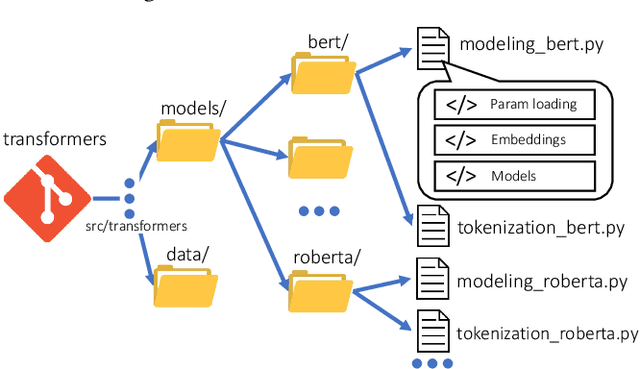
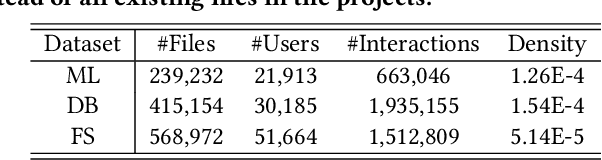
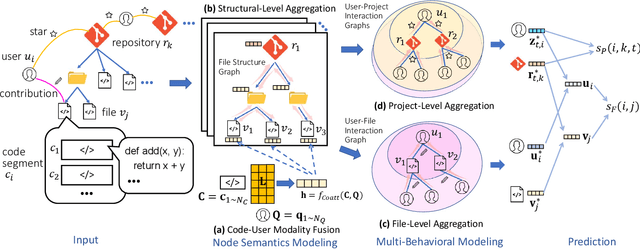
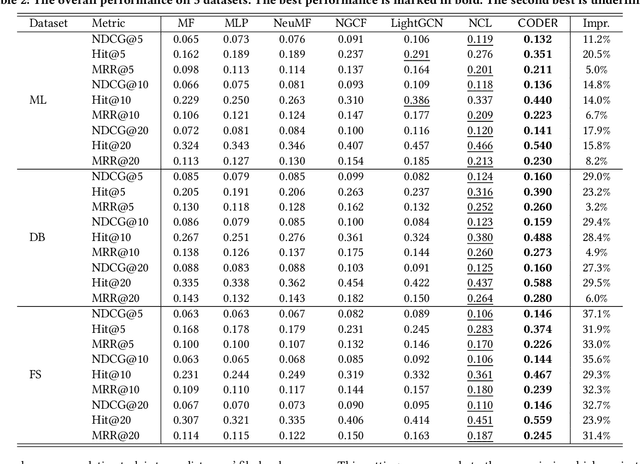
Abstract:Open Source Software (OSS) is forming the spines of technology infrastructures, attracting millions of talents to contribute. Notably, it is challenging and critical to consider both the developers' interests and the semantic features of the project code to recommend appropriate development tasks to OSS developers. In this paper, we formulate the novel problem of code recommendation, whose purpose is to predict the future contribution behaviors of developers given their interaction history, the semantic features of source code, and the hierarchical file structures of projects. Considering the complex interactions among multiple parties within the system, we propose CODER, a novel graph-based code recommendation framework for open source software developers. CODER jointly models microscopic user-code interactions and macroscopic user-project interactions via a heterogeneous graph and further bridges the two levels of information through aggregation on file-structure graphs that reflect the project hierarchy. Moreover, due to the lack of reliable benchmarks, we construct three large-scale datasets to facilitate future research in this direction. Extensive experiments show that our CODER framework achieves superior performance under various experimental settings, including intra-project, cross-project, and cold-start recommendation. We will release all the datasets, code, and utilities for data retrieval upon the acceptance of this work.
Dual-Geometric Space Embedding Model for Two-View Knowledge Graphs
Sep 19, 2022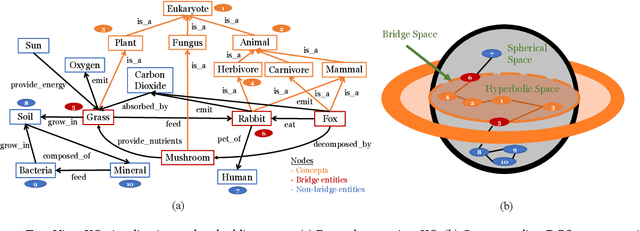
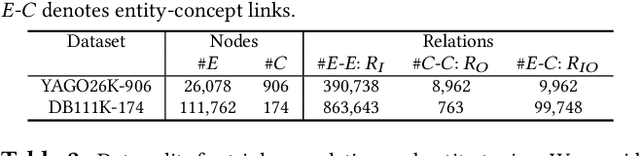
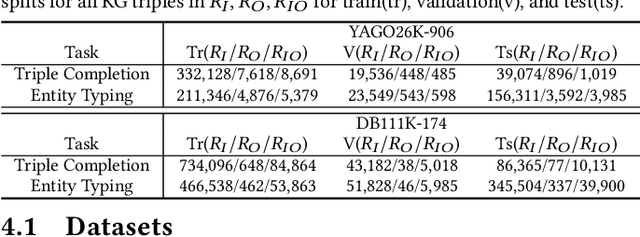
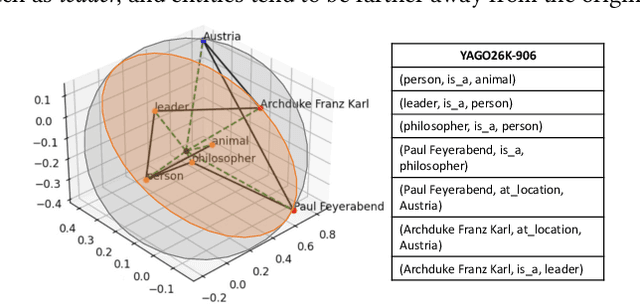
Abstract:Two-view knowledge graphs (KGs) jointly represent two components: an ontology view for abstract and commonsense concepts, and an instance view for specific entities that are instantiated from ontological concepts. As such, these KGs contain heterogeneous structures that are hierarchical, from the ontology-view, and cyclical, from the instance-view. Despite these various structures in KGs, most recent works on embedding KGs assume that the entire KG belongs to only one of the two views but not both simultaneously. For works that seek to put both views of the KG together, the instance and ontology views are assumed to belong to the same geometric space, such as all nodes embedded in the same Euclidean space or non-Euclidean product space, an assumption no longer reasonable for two-view KGs where different portions of the graph exhibit different structures. To address this issue, we define and construct a dual-geometric space embedding model (DGS) that models two-view KGs using a complex non-Euclidean geometric space, by embedding different portions of the KG in different geometric spaces. DGS utilizes the spherical space, hyperbolic space, and their intersecting space in a unified framework for learning embeddings. Furthermore, for the spherical space, we propose novel closed spherical space operators that directly operate in the spherical space without the need for mapping to an approximate tangent space. Experiments on public datasets show that DGS significantly outperforms previous state-of-the-art baseline models on KG completion tasks, demonstrating its ability to better model heterogeneous structures in KGs.
Subgraph Matching via Query-Conditioned Subgraph Matching Neural Networks and Bi-Level Tree Search
Jul 21, 2022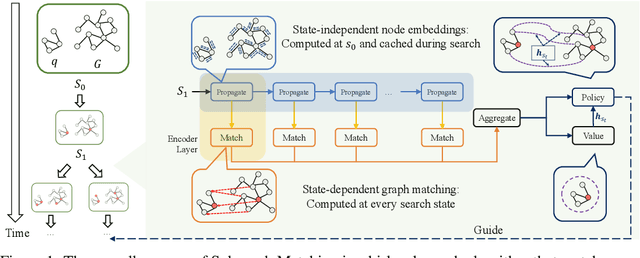
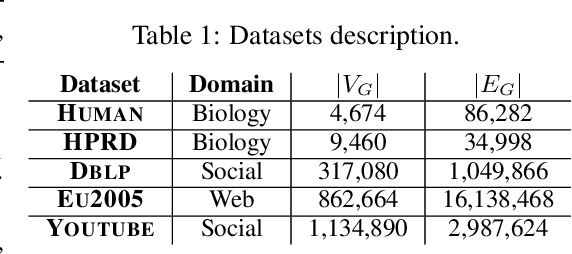
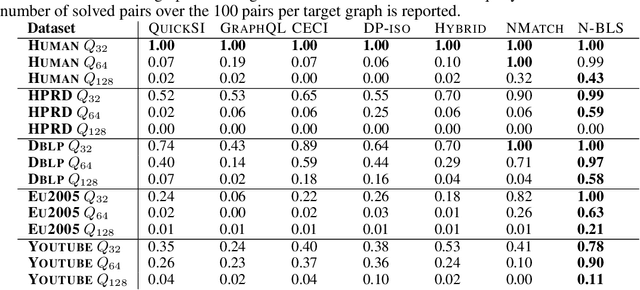
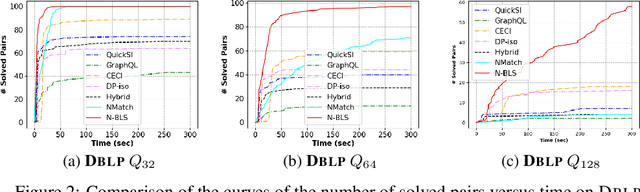
Abstract:Recent advances have shown the success of using reinforcement learning and search to solve NP-hard graph-related tasks, such as Traveling Salesman Optimization, Graph Edit Distance computation, etc. However, it remains unclear how one can efficiently and accurately detect the occurrences of a small query graph in a large target graph, which is a core operation in graph database search, biomedical analysis, social group finding, etc. This task is called Subgraph Matching which essentially performs subgraph isomorphism check between a query graph and a large target graph. One promising approach to this classical problem is the "learning-to-search" paradigm, where a reinforcement learning (RL) agent is designed with a learned policy to guide a search algorithm to quickly find the solution without any solved instances for supervision. However, for the specific task of Subgraph Matching, though the query graph is usually small given by the user as input, the target graph is often orders-of-magnitude larger. It poses challenges to the neural network design and can lead to solution and reward sparsity. In this paper, we propose N-BLS with two innovations to tackle the challenges: (1) A novel encoder-decoder neural network architecture to dynamically compute the matching information between the query and the target graphs at each search state; (2) A Monte Carlo Tree Search enhanced bi-level search framework for training the policy and value networks. Experiments on five large real-world target graphs show that N-BLS can significantly improve the subgraph matching performance.
Enabling Automated FPGA Accelerator Optimization Using Graph Neural Networks
Nov 22, 2021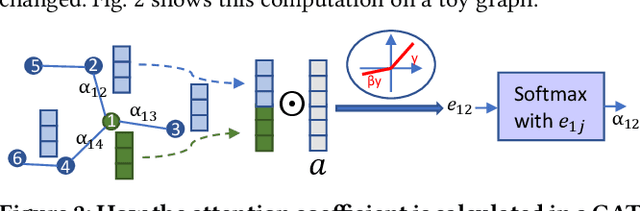
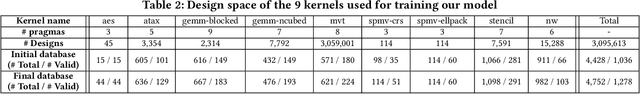
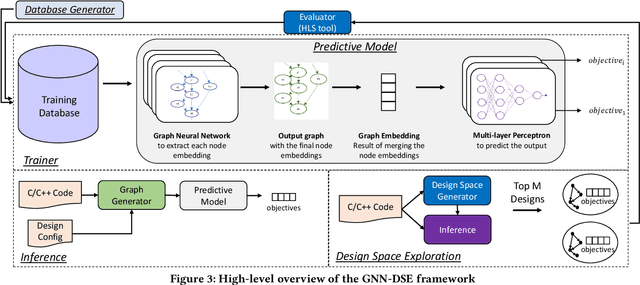
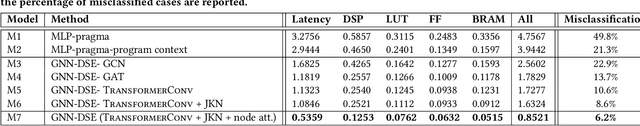
Abstract:High-level synthesis (HLS) has freed the computer architects from developing their designs in a very low-level language and needing to exactly specify how the data should be transferred in register-level. With the help of HLS, the hardware designers must describe only a high-level behavioral flow of the design. Despite this, it still can take weeks to develop a high-performance architecture mainly because there are many design choices at a higher level that requires more time to explore. It also takes several minutes to hours to get feedback from the HLS tool on the quality of each design candidate. In this paper, we propose to solve this problem by modeling the HLS tool with a graph neural network (GNN) that is trained to be used for a wide range of applications. The experimental results demonstrate that by employing the GNN-based model, we are able to estimate the quality of design in milliseconds with high accuracy which can help us search through the solution space very quickly.
Multivariate Time Series Classification with Hierarchical Variational Graph Pooling
Oct 12, 2020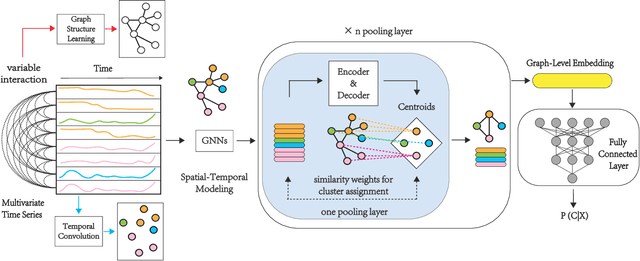
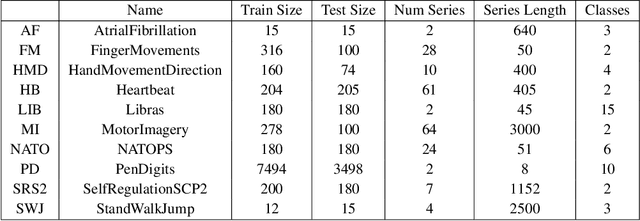
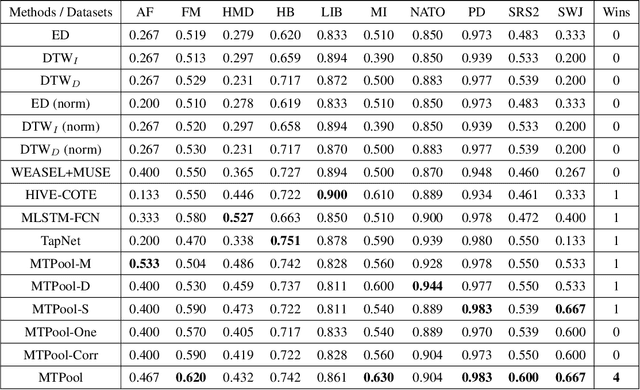
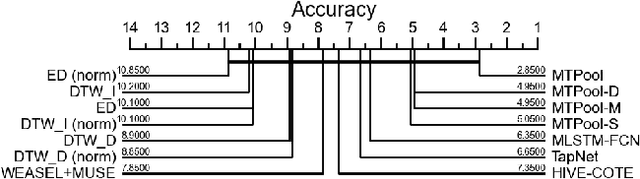
Abstract:Over the past decade, multivariate time series classification (MTSC) has received great attention with the advance of sensing techniques. Current deep learning methods for MTSC are based on convolutional and recurrent neural network, with the assumption that time series variables have the same effect to each other. Thus they cannot model the pairwise dependencies among variables explicitly. What's more, current spatial-temporal modeling methods based on GNNs are inherently flat and lack the capability of aggregating node information in a hierarchical manner. To address this limitation and attain expressive global representation of MTS, we propose a graph pooling based framework MTPool and view MTSC task as graph classification task. With graph structure learning and temporal convolution, MTS slices are converted to graphs and spatial-temporal features are extracted. Then, we propose a novel graph pooling method, which uses an ``encoder-decoder'' mechanism to generate adaptive centroids for cluster assignments. GNNs and graph pooling layers are used for joint graph representation learning and graph coarsening. With multiple graph pooling layers, the input graphs are hierachically coarsened to one node. Finally, differentiable classifier takes this coarsened one-node graph as input to get the final predicted class. Experiments on 10 benchmark datasets demonstrate MTPool outperforms state-of-the-art methods in MTSC tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge