Yufang Huang
AutoShot: A Short Video Dataset and State-of-the-Art Shot Boundary Detection
Apr 12, 2023Abstract:The short-form videos have explosive popularity and have dominated the new social media trends. Prevailing short-video platforms,~\textit{e.g.}, Kuaishou (Kwai), TikTok, Instagram Reels, and YouTube Shorts, have changed the way we consume and create content. For video content creation and understanding, the shot boundary detection (SBD) is one of the most essential components in various scenarios. In this work, we release a new public Short video sHot bOundary deTection dataset, named SHOT, consisting of 853 complete short videos and 11,606 shot annotations, with 2,716 high quality shot boundary annotations in 200 test videos. Leveraging this new data wealth, we propose to optimize the model design for video SBD, by conducting neural architecture search in a search space encapsulating various advanced 3D ConvNets and Transformers. Our proposed approach, named AutoShot, achieves higher F1 scores than previous state-of-the-art approaches, e.g., outperforming TransNetV2 by 4.2%, when being derived and evaluated on our newly constructed SHOT dataset. Moreover, to validate the generalizability of the AutoShot architecture, we directly evaluate it on another three public datasets: ClipShots, BBC and RAI, and the F1 scores of AutoShot outperform previous state-of-the-art approaches by 1.1%, 0.9% and 1.2%, respectively. The SHOT dataset and code can be found in https://github.com/wentaozhu/AutoShot.git .
Test-Time Training for Deformable Multi-Scale Image Registration
Mar 25, 2021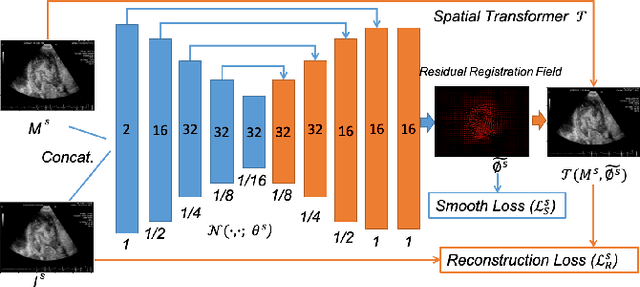
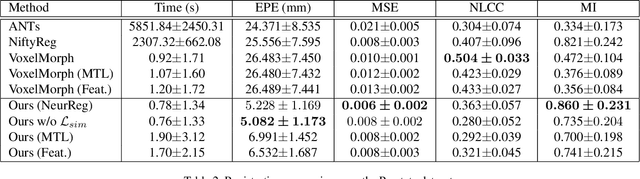
Abstract:Registration is a fundamental task in medical robotics and is often a crucial step for many downstream tasks such as motion analysis, intra-operative tracking and image segmentation. Popular registration methods such as ANTs and NiftyReg optimize objective functions for each pair of images from scratch, which are time-consuming for 3D and sequential images with complex deformations. Recently, deep learning-based registration approaches such as VoxelMorph have been emerging and achieve competitive performance. In this work, we construct a test-time training for deep deformable image registration to improve the generalization ability of conventional learning-based registration model. We design multi-scale deep networks to consecutively model the residual deformations, which is effective for high variational deformations. Extensive experiments validate the effectiveness of multi-scale deep registration with test-time training based on Dice coefficient for image segmentation and mean square error (MSE), normalized local cross-correlation (NLCC) for tissue dense tracking tasks. Two videos are in https://www.youtube.com/watch?v=NvLrCaqCiAE and https://www.youtube.com/watch?v=pEA6ZmtTNuQ
* ICRA 2021; 8 pages, 4 figures, 2 big tables
DICE: Deep Significance Clustering for Outcome-Aware Stratification
Jan 07, 2021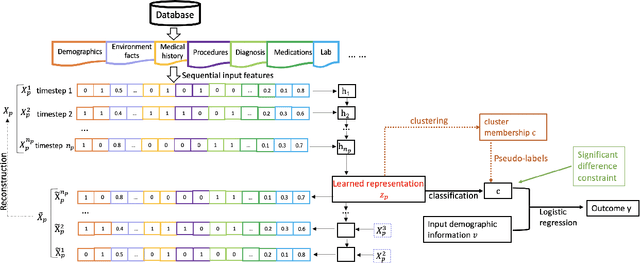
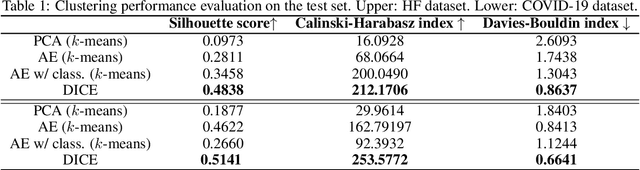
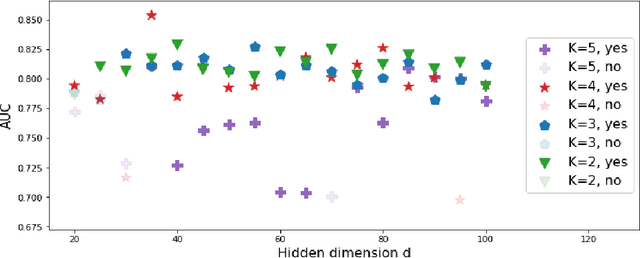
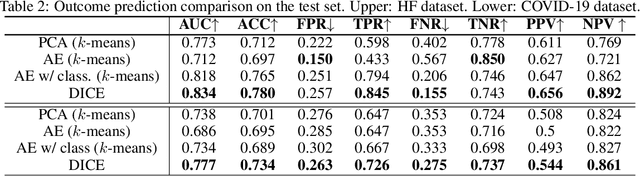
Abstract:We present deep significance clustering (DICE), a framework for jointly performing representation learning and clustering for "outcome-aware" stratification. DICE is intended to generate cluster membership that may be used to categorize a population by individual risk level for a targeted outcome. Following the representation learning and clustering steps, we embed the objective function in DICE with a constraint which requires a statistically significant association between the outcome and cluster membership of learned representations. DICE further includes a neural architecture search step to maximize both the likelihood of representation learning and outcome classification accuracy with cluster membership as the predictor. To demonstrate its utility in medicine for patient risk-stratification, the performance of DICE was evaluated using two datasets with different outcome ratios extracted from real-world electronic health records. Outcomes are defined as acute kidney injury (30.4\%) among a cohort of COVID-19 patients, and discharge disposition (36.8\%) among a cohort of heart failure patients, respectively. Extensive results demonstrate that DICE has superior performance as measured by the difference in outcome distribution across clusters, Silhouette score, Calinski-Harabasz index, and Davies-Bouldin index for clustering, and Area under the ROC Curve (AUC) for outcome classification compared to several baseline approaches.
Cycle-Consistent Adversarial Autoencoders for Unsupervised Text Style Transfer
Oct 02, 2020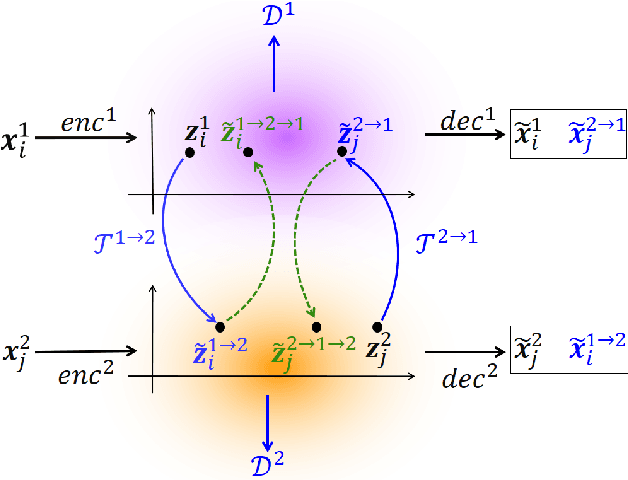

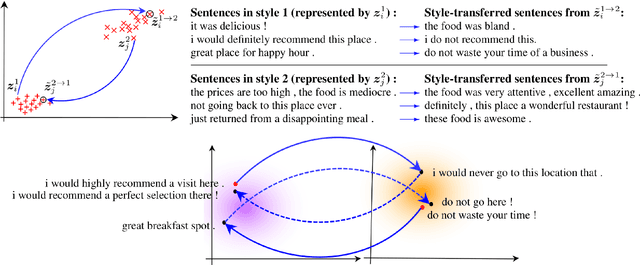
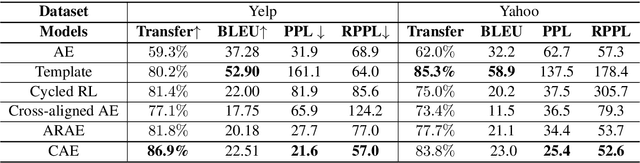
Abstract:Unsupervised text style transfer is full of challenges due to the lack of parallel data and difficulties in content preservation. In this paper, we propose a novel neural approach to unsupervised text style transfer, which we refer to as Cycle-consistent Adversarial autoEncoders (CAE) trained from non-parallel data. CAE consists of three essential components: (1) LSTM autoencoders that encode a text in one style into its latent representation and decode an encoded representation into its original text or a transferred representation into a style-transferred text, (2) adversarial style transfer networks that use an adversarially trained generator to transform a latent representation in one style into a representation in another style, and (3) a cycle-consistent constraint that enhances the capacity of the adversarial style transfer networks in content preservation. The entire CAE with these three components can be trained end-to-end. Extensive experiments and in-depth analyses on two widely-used public datasets consistently validate the effectiveness of proposed CAE in both style transfer and content preservation against several strong baselines in terms of four automatic evaluation metrics and human evaluation.
NeurReg: Neural Registration and Its Application to Image Segmentation
Oct 04, 2019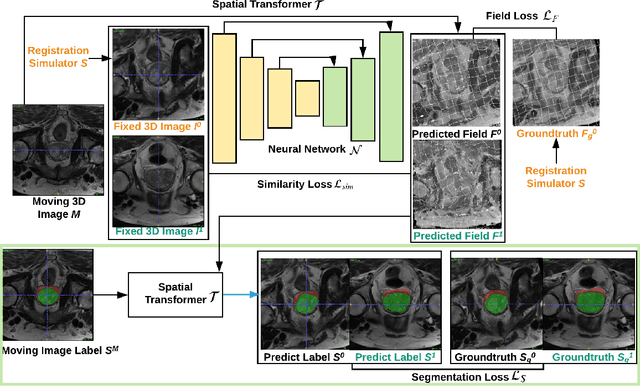


Abstract:Registration is a fundamental task in medical image analysis which can be applied to several tasks including image segmentation, intra-operative tracking, multi-modal image alignment, and motion analysis. Popular registration tools such as ANTs and NiftyReg optimize an objective function for each pair of images from scratch which is time-consuming for large images with complicated deformation. Facilitated by the rapid progress of deep learning, learning-based approaches such as VoxelMorph have been emerging for image registration. These approaches can achieve competitive performance in a fraction of a second on advanced GPUs. In this work, we construct a neural registration framework, called NeurReg, with a hybrid loss of displacement fields and data similarity, which substantially improves the current state-of-the-art of registrations. Within the framework, we simulate various transformations by a registration simulator which generates fixed image and displacement field ground truth for training. Furthermore, we design three segmentation frameworks based on the proposed registration framework: 1) atlas-based segmentation, 2) joint learning of both segmentation and registration tasks, and 3) multi-task learning with atlas-based segmentation as an intermediate feature. Extensive experimental results validate the effectiveness of the proposed NeurReg framework based on various metrics: the endpoint error (EPE) of the predicted displacement field, mean square error (MSE), normalized local cross-correlation (NLCC), mutual information (MI), Dice coefficient, uncertainty estimation, and the interpretability of the segmentation. The proposed NeurReg improves registration accuracy with fast inference speed, which can greatly accelerate related medical image analysis tasks.
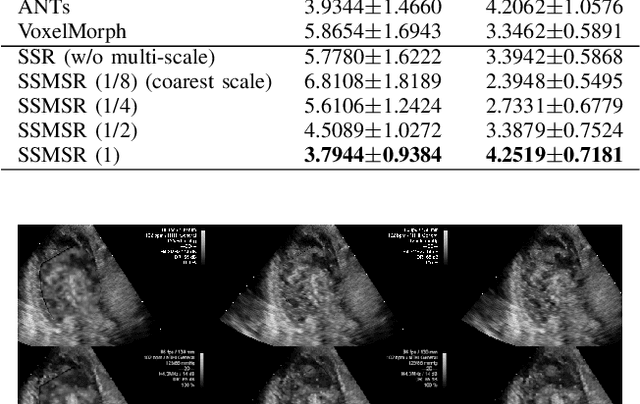
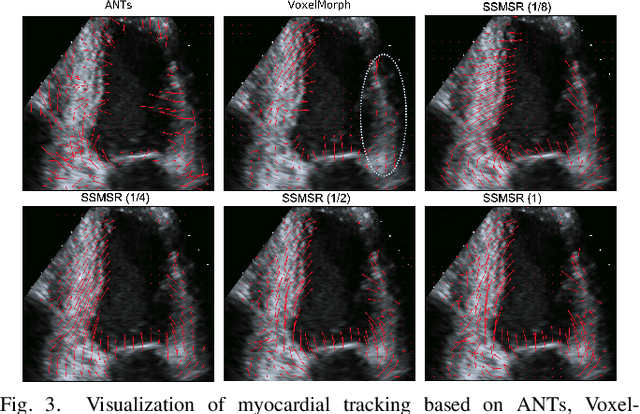
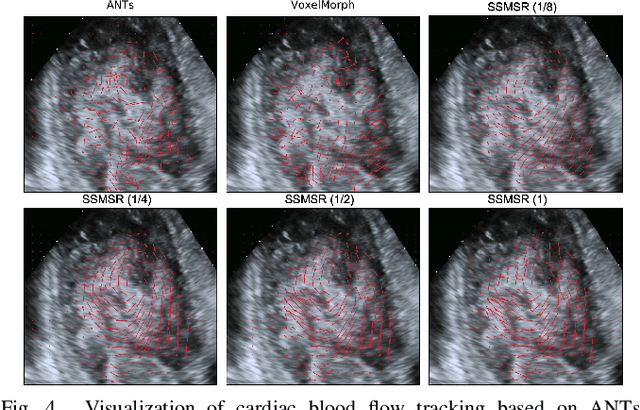
Neural Multi-Scale Self-Supervised Registration for Echocardiogram Dense Tracking
Jun 18, 2019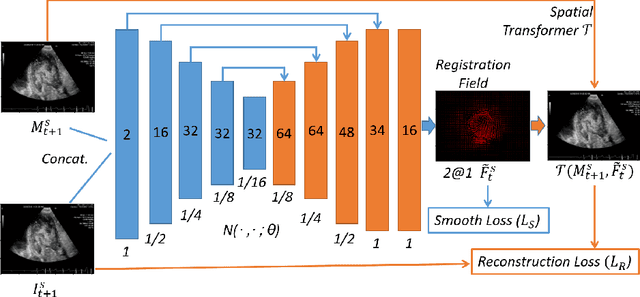
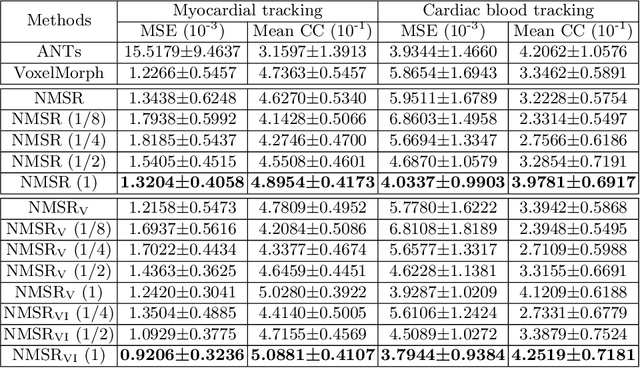
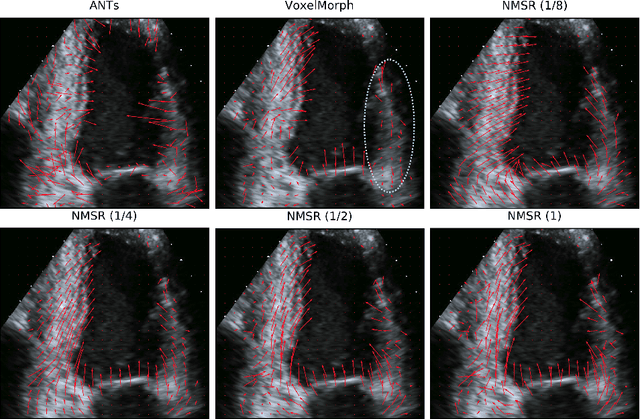
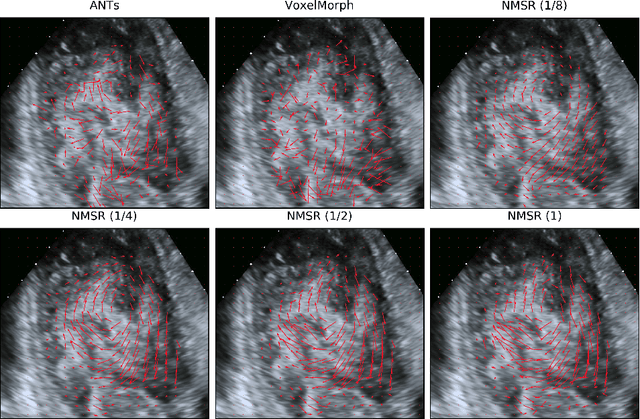
Abstract:Echocardiography has become routinely used in the diagnosis of cardiomyopathy and abnormal cardiac blood flow. However, manually measuring myocardial motion and cardiac blood flow from echocardiogram is time-consuming and error-prone. Computer algorithms that can automatically track and quantify myocardial motion and cardiac blood flow are highly sought after, but have not been very successful due to noise and high variability of echocardiography. In this work, we propose a neural multi-scale self-supervised registration (NMSR) method for automated myocardial and cardiac blood flow dense tracking. NMSR incorporates two novel components: 1) utilizing a deep neural net to parameterize the velocity field between two image frames, and 2) optimizing the parameters of the neural net in a sequential multi-scale fashion to account for large variations within the velocity field. Experiments demonstrate that NMSR yields significantly better registration accuracy than state-of-the-art methods, such as advanced normalization tools (ANTs) and VoxelMorph, for both myocardial and cardiac blood flow dense tracking. Our approach promises to provide a fully automated method for fast and accurate analyses of echocardiograms.
AnatomyNet: Deep 3D Squeeze-and-excitation U-Nets for fast and fully automated whole-volume anatomical segmentation
Aug 15, 2018



Abstract:Radiation therapy (RT) is a common treatment for head and neck (HaN) cancer where therapists are often required to manually delineate boundaries of the organs-at-risks (OARs). Automated head and neck anatomical segmentation provides a way to speed up and improve the reproducibility of radiation therapy planning. In this work, we propose the AnatomyNet, an end-to-end and atlas-free three dimensional squeeze-and-excitation U-Net (3D SE U-Net), for fast and fully automated whole-volume HaN anatomical segmentation. There are two main challenges for fully automated HaN OARs segmentation: 1) challenge in segmenting small anatomies (i.e., optic chiasm and optic nerves) occupying only a few slices, and 2) training model with inconsistent data annotations with missing ground truth for some anatomical structures because of different RT planning. We propose the AnatomyNet that has one down-sampling layer with the trade-off between GPU memory and feature representation capacity, and 3D SE residual blocks for effective feature learning to alleviate these challenges. Moreover, we design a hybrid loss function with the Dice loss and the focal loss. The Dice loss is a class level distribution loss that depends less on the number of voxels in the anatomy, and the focal loss is designed to deal with highly unbalanced segmentation. For missing annotations, we propose masked loss and weighted loss for accurate and balanced weights updating in the learning of the AnatomyNet. We collect 261 HaN CT images to train the AnatomyNet for segmenting nine anatomies. Compared to previous state-of-the-art methods for each anatomy from the MICCAI 2015 competition, the AnatomyNet increases Dice similarity coefficient (DSC) by 3.3% on average. The proposed AnatomyNet takes only 0.12 seconds on average to segment a whole-volume HaN CT image of an average dimension of 178x302x225.
DeepEM: Deep 3D ConvNets With EM For Weakly Supervised Pulmonary Nodule Detection
May 26, 2018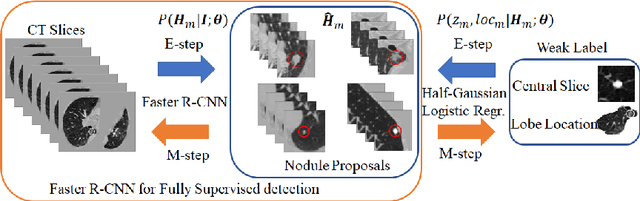
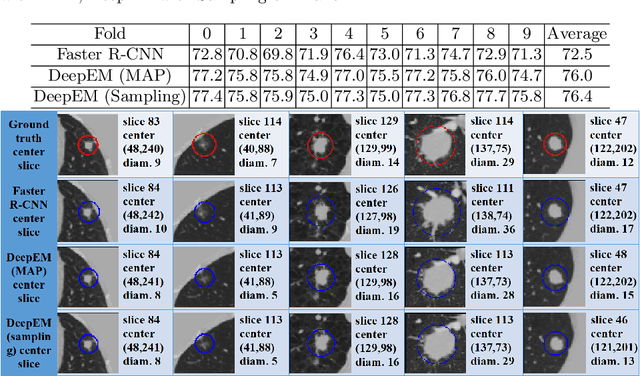
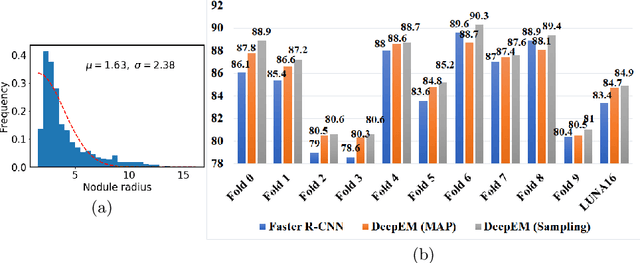
Abstract:Recently deep learning has been witnessing widespread adoption in various medical image applications. However, training complex deep neural nets requires large-scale datasets labeled with ground truth, which are often unavailable in many medical image domains. For instance, to train a deep neural net to detect pulmonary nodules in lung computed tomography (CT) images, current practice is to manually label nodule locations and sizes in many CT images to construct a sufficiently large training dataset, which is costly and difficult to scale. On the other hand, electronic medical records (EMR) contain plenty of partial information on the content of each medical image. In this work, we explore how to tap this vast, but currently unexplored data source to improve pulmonary nodule detection. We propose DeepEM, a novel deep 3D ConvNet framework augmented with expectation-maximization (EM), to mine weakly supervised labels in EMRs for pulmonary nodule detection. Experimental results show that DeepEM can lead to 1.5\% and 3.9\% average improvement in free-response receiver operating characteristic (FROC) scores on LUNA16 and Tianchi datasets, respectively, demonstrating the utility of incomplete information in EMRs for improving deep learning algorithms.\footnote{https://github.com/uci-cbcl/DeepEM-for-Weakly-Supervised-Detection.git}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge