Yili Zhao
Habitat-Matterport 3D Dataset (HM3D): 1000 Large-scale 3D Environments for Embodied AI
Sep 16, 2021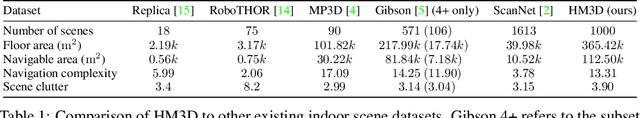

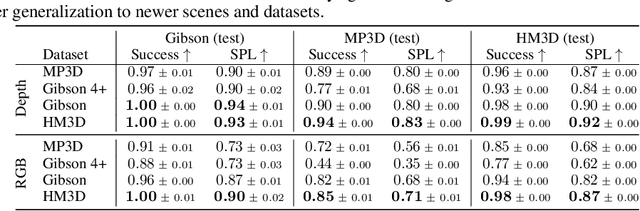
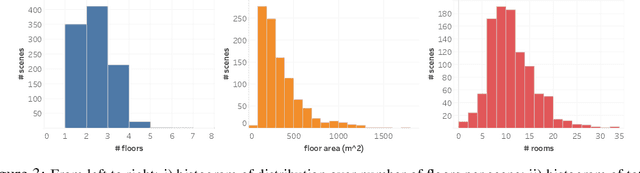
Abstract:We present the Habitat-Matterport 3D (HM3D) dataset. HM3D is a large-scale dataset of 1,000 building-scale 3D reconstructions from a diverse set of real-world locations. Each scene in the dataset consists of a textured 3D mesh reconstruction of interiors such as multi-floor residences, stores, and other private indoor spaces. HM3D surpasses existing datasets available for academic research in terms of physical scale, completeness of the reconstruction, and visual fidelity. HM3D contains 112.5k m^2 of navigable space, which is 1.4 - 3.7x larger than other building-scale datasets such as MP3D and Gibson. When compared to existing photorealistic 3D datasets such as Replica, MP3D, Gibson, and ScanNet, images rendered from HM3D have 20 - 85% higher visual fidelity w.r.t. counterpart images captured with real cameras, and HM3D meshes have 34 - 91% fewer artifacts due to incomplete surface reconstruction. The increased scale, fidelity, and diversity of HM3D directly impacts the performance of embodied AI agents trained using it. In fact, we find that HM3D is `pareto optimal' in the following sense -- agents trained to perform PointGoal navigation on HM3D achieve the highest performance regardless of whether they are evaluated on HM3D, Gibson, or MP3D. No similar claim can be made about training on other datasets. HM3D-trained PointNav agents achieve 100% performance on Gibson-test dataset, suggesting that it might be time to retire that episode dataset.
Habitat 2.0: Training Home Assistants to Rearrange their Habitat
Jun 28, 2021
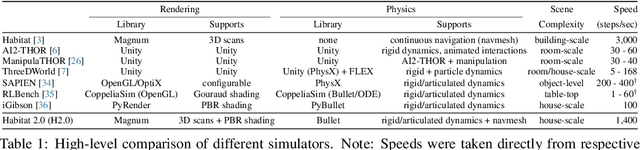

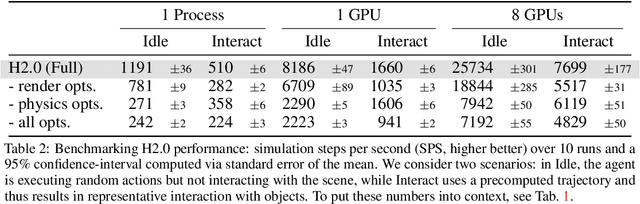
Abstract:We introduce Habitat 2.0 (H2.0), a simulation platform for training virtual robots in interactive 3D environments and complex physics-enabled scenarios. We make comprehensive contributions to all levels of the embodied AI stack - data, simulation, and benchmark tasks. Specifically, we present: (i) ReplicaCAD: an artist-authored, annotated, reconfigurable 3D dataset of apartments (matching real spaces) with articulated objects (e.g. cabinets and drawers that can open/close); (ii) H2.0: a high-performance physics-enabled 3D simulator with speeds exceeding 25,000 simulation steps per second (850x real-time) on an 8-GPU node, representing 100x speed-ups over prior work; and, (iii) Home Assistant Benchmark (HAB): a suite of common tasks for assistive robots (tidy the house, prepare groceries, set the table) that test a range of mobile manipulation capabilities. These large-scale engineering contributions allow us to systematically compare deep reinforcement learning (RL) at scale and classical sense-plan-act (SPA) pipelines in long-horizon structured tasks, with an emphasis on generalization to new objects, receptacles, and layouts. We find that (1) flat RL policies struggle on HAB compared to hierarchical ones; (2) a hierarchy with independent skills suffers from 'hand-off problems', and (3) SPA pipelines are more brittle than RL policies.
Habitat: A Platform for Embodied AI Research
Apr 02, 2019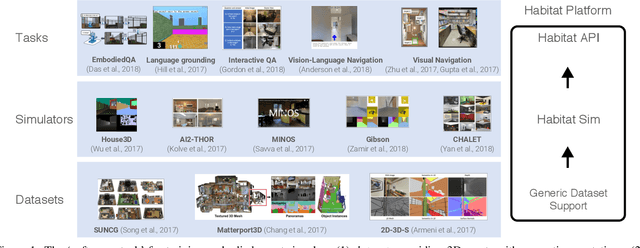
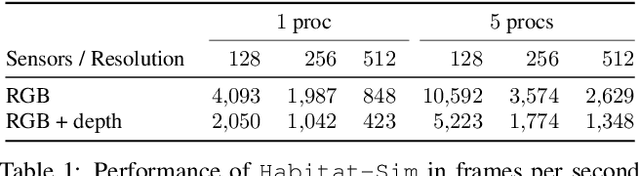

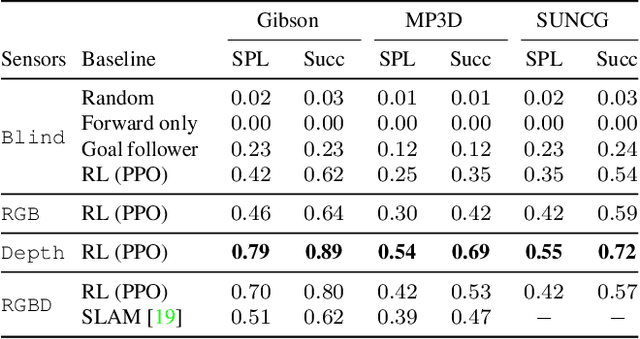
Abstract:We present Habitat, a new platform for research in embodied artificial intelligence (AI). Habitat enables training embodied agents (virtual robots) in highly efficient photorealistic 3D simulation, before transferring the learned skills to reality. Specifically, Habitat consists of the following: 1. Habitat-Sim: a flexible, high-performance 3D simulator with configurable agents, multiple sensors, and generic 3D dataset handling (with built-in support for SUNCG, Matterport3D, Gibson datasets). Habitat-Sim is fast -- when rendering a scene from the Matterport3D dataset, Habitat-Sim achieves several thousand frames per second (fps) running single-threaded, and can reach over 10,000 fps multi-process on a single GPU, which is orders of magnitude faster than the closest simulator. 2. Habitat-API: a modular high-level library for end-to-end development of embodied AI algorithms -- defining embodied AI tasks (e.g. navigation, instruction following, question answering), configuring and training embodied agents (via imitation or reinforcement learning, or via classic SLAM), and benchmarking using standard metrics. These large-scale engineering contributions enable us to answer scientific questions requiring experiments that were till now impracticable or `merely' impractical. Specifically, in the context of point-goal navigation (1) we revisit the comparison between learning and SLAM approaches from two recent works and find evidence for the opposite conclusion -- that learning outperforms SLAM, if scaled to total experience far surpassing that of previous investigations, and (2) we conduct the first cross-dataset generalization experiments {train, test} x {Matterport3D, Gibson} for multiple sensors {blind, RGB, RGBD, D} and find that only agents with depth (D) sensors generalize across datasets. We hope that our open-source platform and these findings will advance research in embodied AI.
Deep-learning models improve on community-level diagnosis for common congenital heart disease lesions
Sep 19, 2018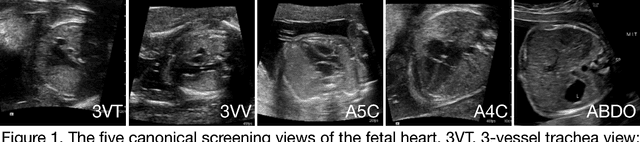
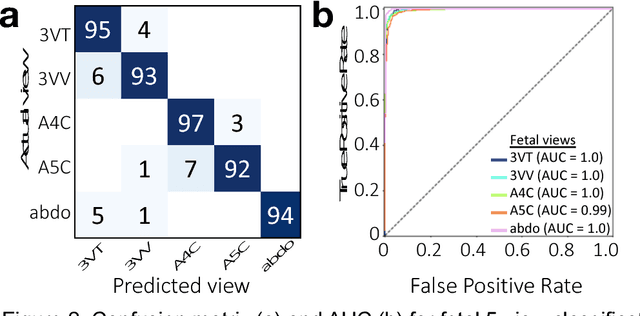

Abstract:Prenatal diagnosis of tetralogy of Fallot (TOF) and hypoplastic left heart syndrome (HLHS), two serious congenital heart defects, improves outcomes and can in some cases facilitate in utero interventions. In practice, however, the fetal diagnosis rate for these lesions is only 30-50 percent in community settings. Improving fetal diagnosis of congenital heart disease is therefore critical. Deep learning is a cutting-edge machine learning technique for finding patterns in images but has not yet been applied to prenatal diagnosis of congenital heart disease. Using 685 retrospectively collected echocardiograms from fetuses 18-24 weeks of gestational age from 2000-2018, we trained convolutional and fully-convolutional deep learning models in a supervised manner to (i) identify the five canonical screening views of the fetal heart and (ii) segment cardiac structures to calculate fetal cardiac biometrics. We then trained models to distinguish by view between normal hearts, TOF, and HLHS. In a holdout test set of images, F-score for identification of the five most important fetal cardiac views was 0.95. Binary classification of unannotated cardiac views of normal heart vs. TOF reached an overall sensitivity of 75% and a specificity of 76%, while normal vs. HLHS reached a sensitivity of 100% and specificity of 90%, both well above average diagnostic rates for these lesions. Furthermore, segmentation-based measurements for cardiothoracic ratio (CTR), cardiac axis (CA), and ventricular fractional area change (FAC) were compatible with clinically measured metrics for normal, TOF, and HLHS hearts. Thus, using guideline-recommended imaging, deep learning models can significantly improve detection of fetal congenital heart disease compared to the common standard of care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge