Xinyuan Huang
S3D: A Spatial Steerable Surgical Drilling Framework for Robotic Spinal Fixation Procedures
Jul 02, 2025



Abstract:In this paper, we introduce S3D: A Spatial Steerable Surgical Drilling Framework for Robotic Spinal Fixation Procedures. S3D is designed to enable realistic steerable drilling while accounting for the anatomical constraints associated with vertebral access in spinal fixation (SF) procedures. To achieve this, we first enhanced our previously designed concentric tube Steerable Drilling Robot (CT-SDR) to facilitate steerable drilling across all vertebral levels of the spinal column. Additionally, we propose a four-Phase calibration, registration, and navigation procedure to perform realistic SF procedures on a spine holder phantom by integrating the CT-SDR with a seven-degree-of-freedom robotic manipulator. The functionality of this framework is validated through planar and out-of-plane steerable drilling experiments in vertebral phantoms.
Federated Neural Architecture Search with Model-Agnostic Meta Learning
Apr 08, 2025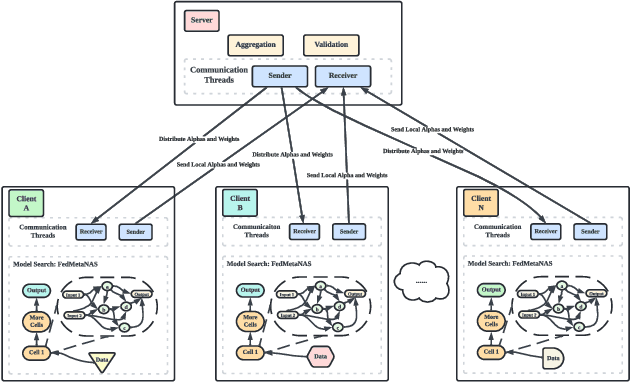
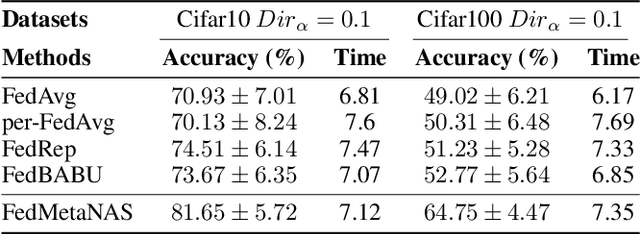
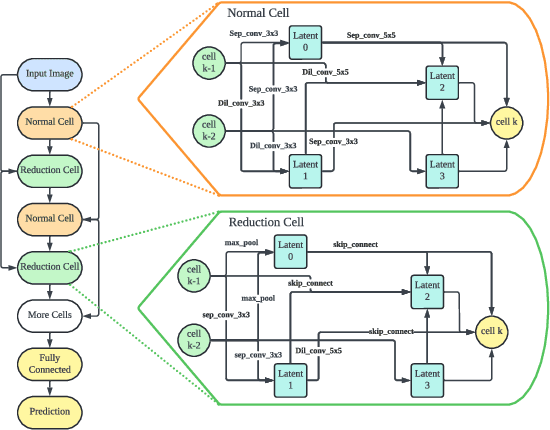
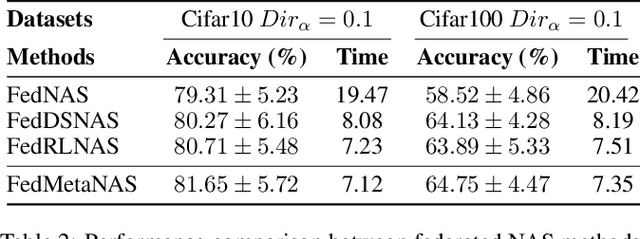
Abstract:Federated Learning (FL) often struggles with data heterogeneity due to the naturally uneven distribution of user data across devices. Federated Neural Architecture Search (NAS) enables collaborative search for optimal model architectures tailored to heterogeneous data to achieve higher accuracy. However, this process is time-consuming due to extensive search space and retraining. To overcome this, we introduce FedMetaNAS, a framework that integrates meta-learning with NAS within the FL context to expedite the architecture search by pruning the search space and eliminating the retraining stage. Our approach first utilizes the Gumbel-Softmax reparameterization to facilitate relaxation of the mixed operations in the search space. We then refine the local search process by incorporating Model-Agnostic Meta-Learning, where a task-specific learner adapts both weights and architecture parameters (alphas) for individual tasks, while a meta learner adjusts the overall model weights and alphas based on the gradient information from task learners. Following the meta-update, we propose soft pruning using the same trick on search space to gradually sparsify the architecture, ensuring that the performance of the chosen architecture remains robust after pruning which allows for immediate use of the model without retraining. Experimental evaluations demonstrate that FedMetaNAS significantly accelerates the search process by more than 50\% with higher accuracy compared to FedNAS.
Independent SE-Equivariant Models for End-to-End Rigid Protein Docking
Nov 15, 2021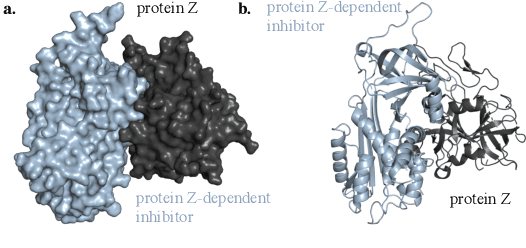
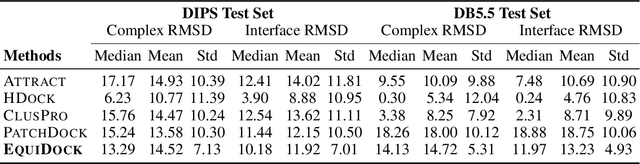
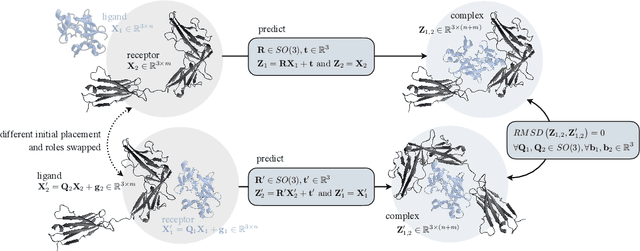

Abstract:Protein complex formation is a central problem in biology, being involved in most of the cell's processes, and essential for applications, e.g. drug design or protein engineering. We tackle rigid body protein-protein docking, i.e., computationally predicting the 3D structure of a protein-protein complex from the individual unbound structures, assuming no conformational change within the proteins happens during binding. We design a novel pairwise-independent SE(3)-equivariant graph matching network to predict the rotation and translation to place one of the proteins at the right docked position relative to the second protein. We mathematically guarantee a basic principle: the predicted complex is always identical regardless of the initial locations and orientations of the two structures. Our model, named EquiDock, approximates the binding pockets and predicts the docking poses using keypoint matching and alignment, achieved through optimal transport and a differentiable Kabsch algorithm. Empirically, we achieve significant running time improvements and often outperform existing docking software despite not relying on heavy candidate sampling, structure refinement, or templates.
MedPerf: Open Benchmarking Platform for Medical Artificial Intelligence using Federated Evaluation
Oct 08, 2021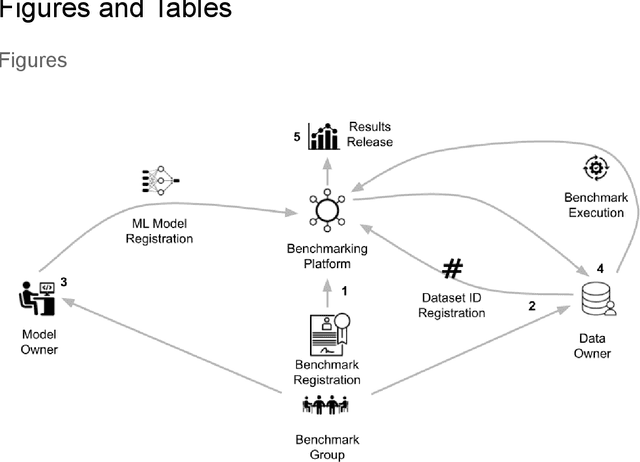
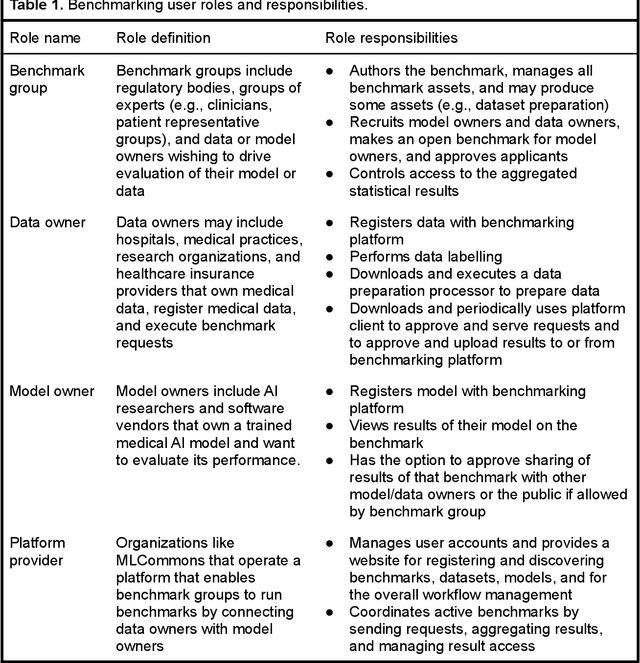
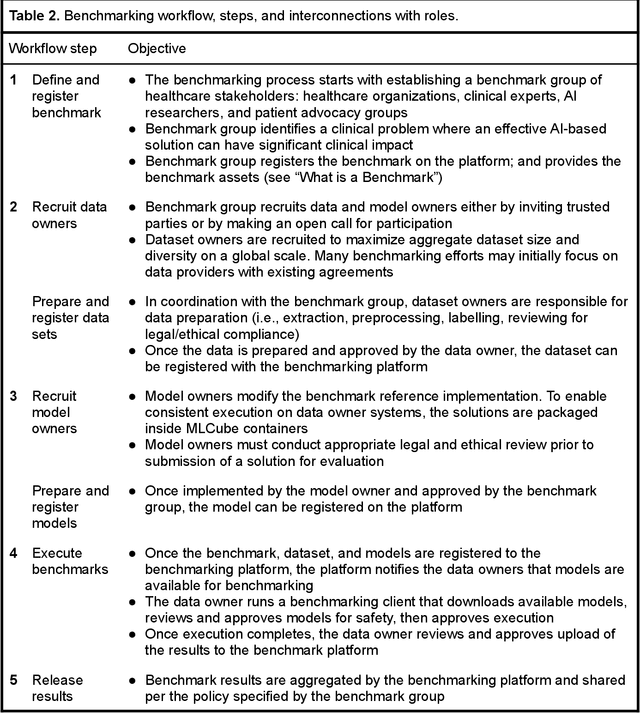
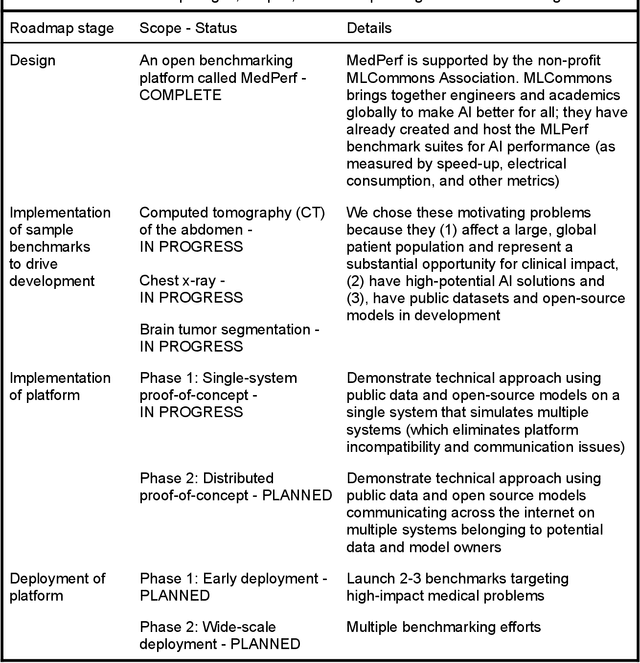
Abstract:Medical AI has tremendous potential to advance healthcare by supporting the evidence-based practice of medicine, personalizing patient treatment, reducing costs, and improving provider and patient experience. We argue that unlocking this potential requires a systematic way to measure the performance of medical AI models on large-scale heterogeneous data. To meet this need, we are building MedPerf, an open framework for benchmarking machine learning in the medical domain. MedPerf will enable federated evaluation in which models are securely distributed to different facilities for evaluation, thereby empowering healthcare organizations to assess and verify the performance of AI models in an efficient and human-supervised process, while prioritizing privacy. We describe the current challenges healthcare and AI communities face, the need for an open platform, the design philosophy of MedPerf, its current implementation status, and our roadmap. We call for researchers and organizations to join us in creating the MedPerf open benchmarking platform.
Benchmarking TinyML Systems: Challenges and Direction
Mar 10, 2020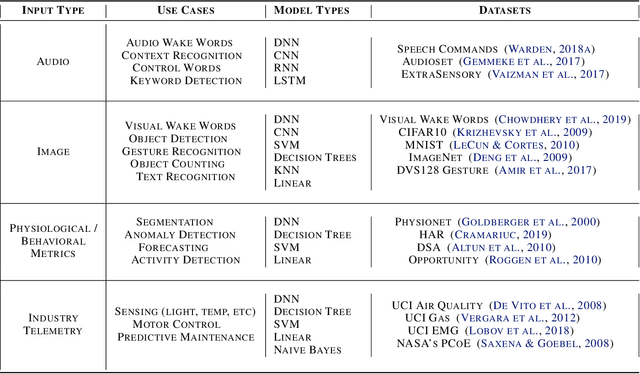
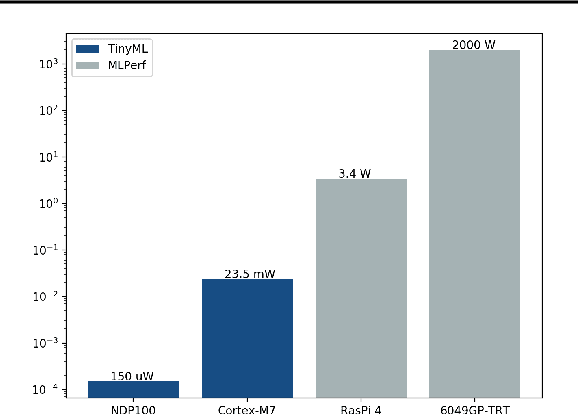
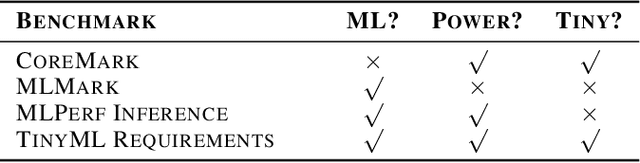
Abstract:Recent advancements in ultra-low-power machine learning (TinyML) hardware promises to unlock an entirely new class of smart applications. However, continued progress is limited by the lack of a widely accepted benchmark for these systems. Benchmarking allows us to measure and thereby systematically compare, evaluate, and improve the performance of systems. In this position paper, we present the current landscape of TinyML and discuss the challenges and direction towards developing a fair and useful hardware benchmark for TinyML workloads. Our viewpoints reflect the collective thoughts of the TinyMLPerf working group that is comprised of 30 organizations.
MLPerf Training Benchmark
Oct 30, 2019
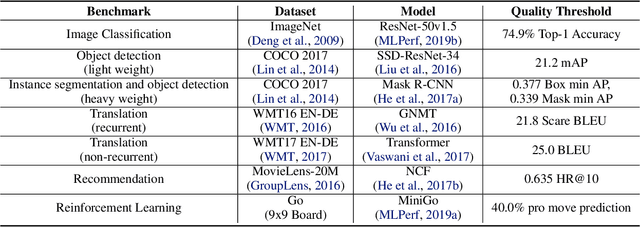
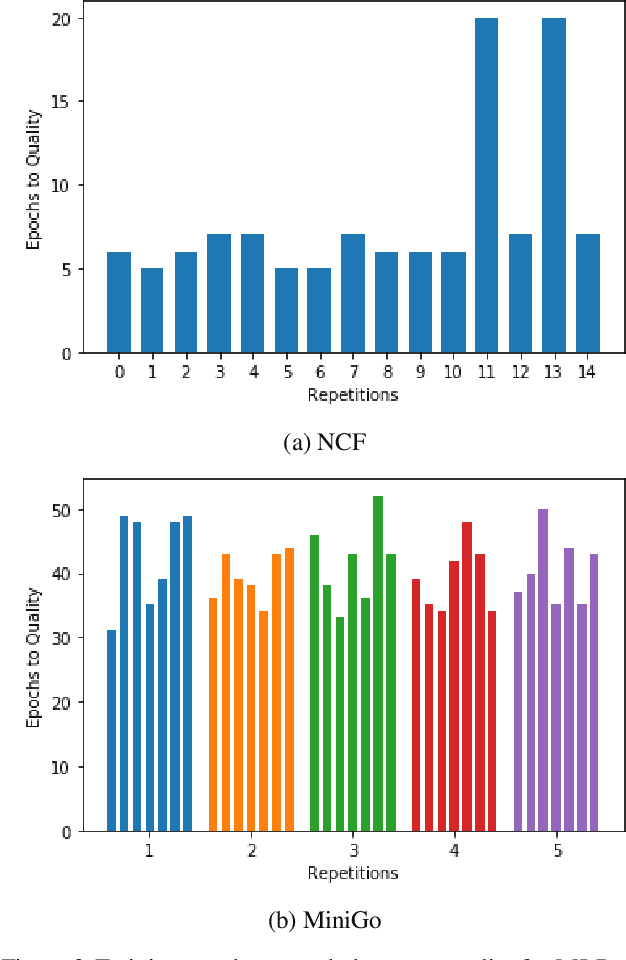
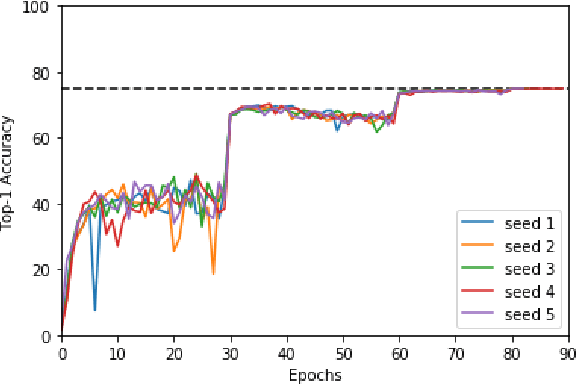
Abstract:Machine learning is experiencing an explosion of software and hardware solutions, and needs industry-standard performance benchmarks to drive design and enable competitive evaluation. However, machine learning training presents a number of unique challenges to benchmarking that do not exist in other domains: (1) some optimizations that improve training throughput actually increase time to solution, (2) training is stochastic and time to solution has high variance, and (3) the software and hardware systems are so diverse that they cannot be fairly benchmarked with the same binary, code, or even hyperparameters. We present MLPerf, a machine learning benchmark that overcomes these challenges. We quantitatively evaluate the efficacy of MLPerf in driving community progress on performance and scalability across two rounds of results from multiple vendors.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge