Xi Long
Advancing Cross-Organ Domain Generalization with Test-Time Style Transfer and Diversity Enhancement
Mar 24, 2025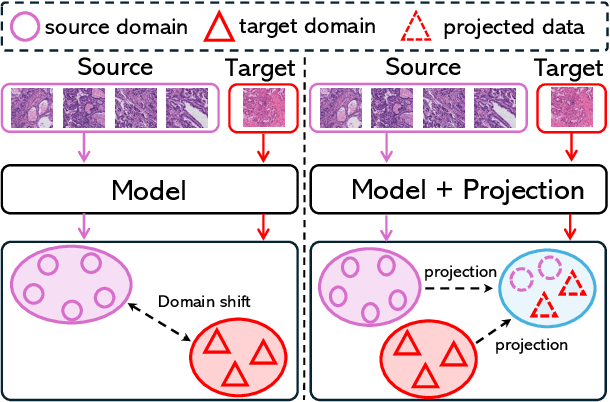



Abstract:Deep learning has made significant progress in addressing challenges in various fields including computational pathology (CPath). However, due to the complexity of the domain shift problem, the performance of existing models will degrade, especially when it comes to multi-domain or cross-domain tasks. In this paper, we propose a Test-time style transfer (T3s) that uses a bidirectional mapping mechanism to project the features of the source and target domains into a unified feature space, enhancing the generalization ability of the model. To further increase the style expression space, we introduce a Cross-domain style diversification module (CSDM) to ensure the orthogonality between style bases. In addition, data augmentation and low-rank adaptation techniques are used to improve feature alignment and sensitivity, enabling the model to adapt to multi-domain inputs effectively. Our method has demonstrated effectiveness on three unseen datasets.
Mitosis domain generalization in histopathology images -- The MIDOG challenge
Apr 06, 2022
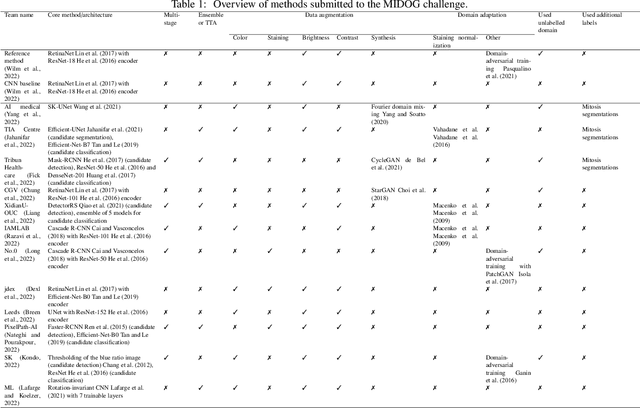
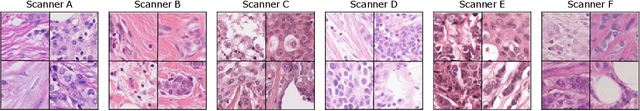
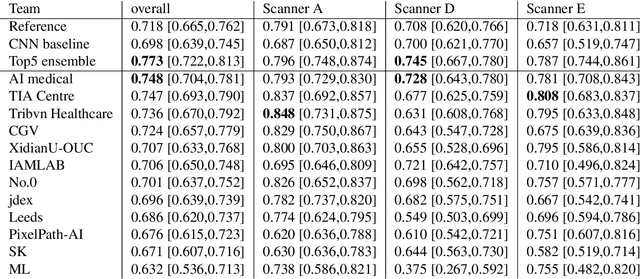
Abstract:The density of mitotic figures within tumor tissue is known to be highly correlated with tumor proliferation and thus is an important marker in tumor grading. Recognition of mitotic figures by pathologists is known to be subject to a strong inter-rater bias, which limits the prognostic value. State-of-the-art deep learning methods can support the expert in this assessment but are known to strongly deteriorate when applied in a different clinical environment than was used for training. One decisive component in the underlying domain shift has been identified as the variability caused by using different whole slide scanners. The goal of the MICCAI MIDOG 2021 challenge has been to propose and evaluate methods that counter this domain shift and derive scanner-agnostic mitosis detection algorithms. The challenge used a training set of 200 cases, split across four scanning systems. As a test set, an additional 100 cases split across four scanning systems, including two previously unseen scanners, were given. The best approaches performed on an expert level, with the winning algorithm yielding an F_1 score of 0.748 (CI95: 0.704-0.781). In this paper, we evaluate and compare the approaches that were submitted to the challenge and identify methodological factors contributing to better performance.
AlphaFold Accelerates Artificial Intelligence Powered Drug Discovery: Efficient Discovery of a Novel Cyclin-dependent Kinase 20 (CDK20) Small Molecule Inhibitor
Jan 21, 2022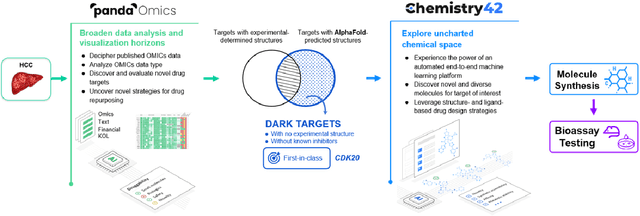
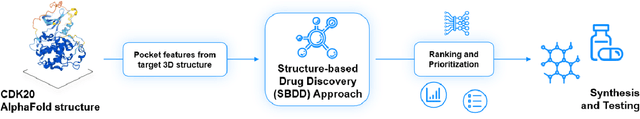
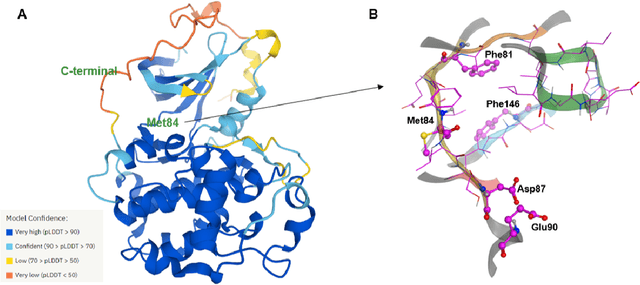
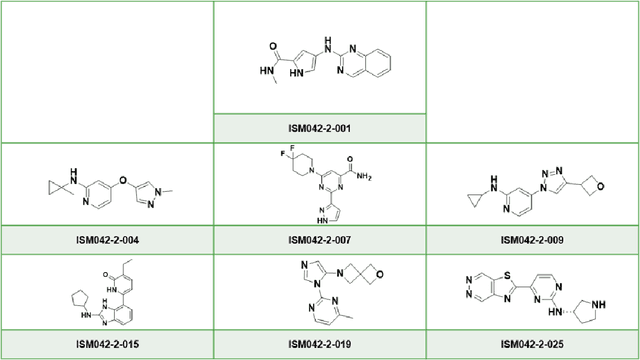
Abstract:The AlphaFold computer program predicted protein structures for the whole human genome, which has been considered as a remarkable breakthrough both in artificial intelligence (AI) application and structural biology. Despite the varying confidence level, these predicted structures still could significantly contribute to the structure-based drug design of novel targets, especially the ones with no or limited structural information. In this work, we successfully applied AlphaFold in our end-to-end AI-powered drug discovery engines constituted of a biocomputational platform PandaOmics and a generative chemistry platform Chemistry42, to identify a first-in-class hit molecule of a novel target without an experimental structure starting from target selection towards hit identification in a cost- and time-efficient manner. PandaOmics provided the targets of interest and Chemistry42 generated the molecules based on the AlphaFold predicted structure, and the selected molecules were synthesized and tested in biological assays. Through this approach, we identified a small molecule hit compound for CDK20 with a Kd value of 8.9 +/- 1.6 uM (n = 4) within 30 days from target selection and after only synthesizing 7 compounds. To the best of our knowledge, this is the first reported small molecule targeting CDK20 and more importantly, this work is the first demonstration of AlphaFold application in the hit identification process in early drug discovery.
Domain Adaptive Cascade R-CNN for MItosis DOmain Generalization Challenge
Sep 29, 2021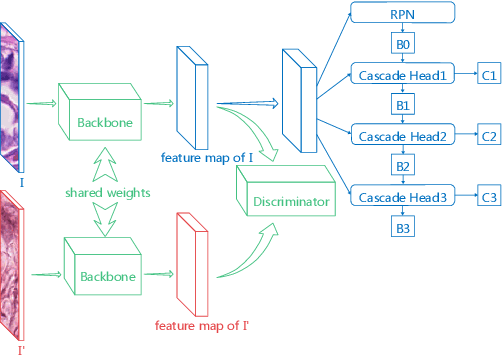
Abstract:We present a summary of the domain adaptive cascade R-CNN method for mitosis detection of digital histopathology images. By comprehensive data augmentation and adapting existing popular detection architecture, our proposed method has achieved an F1 score of 0.7500 on the preliminary test set in MItosis DOmain Generalization (MIDOG) Challenge at MICCAI 2021.
Human Gender Classification: A Review
Mar 16, 2016
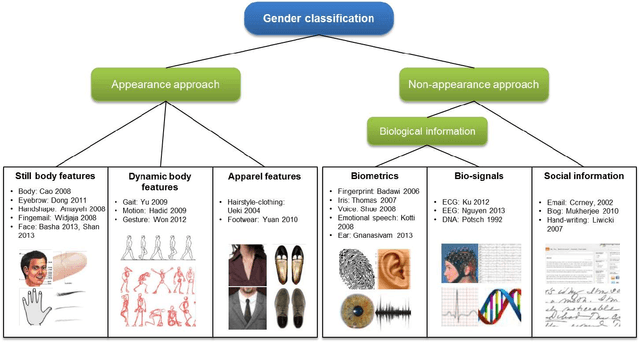
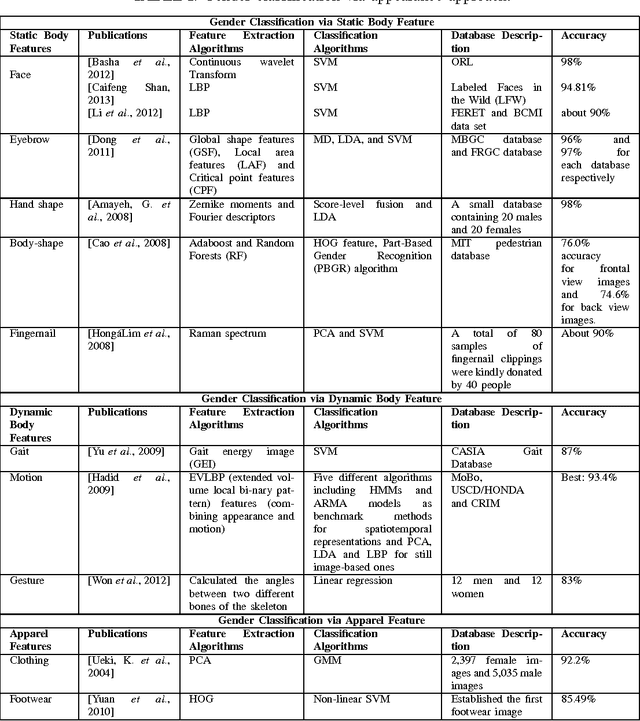
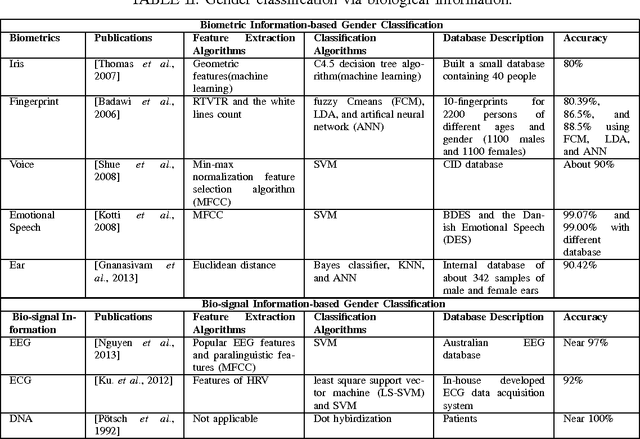
Abstract:Gender contains a wide range of information regarding to the characteristics difference between male and female. Successful gender recognition is essential and critical for many applications in the commercial domains such as applications of human-computer interaction and computer-aided physiological or psychological analysis. Some have proposed various approaches for automatic gender classification using the features derived from human bodies and/or behaviors. First, this paper introduces the challenge and application for gender classification research. Then, the development and framework of gender classification are described. Besides, we compare these state-of-the-art approaches, including vision-based methods, biological information-based method, and social network information-based method, to provide a comprehensive review in the area of gender classification. In mean time, we highlight the strength and discuss the limitation of each method. Finally, this review also discusses several promising applications for the future work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge