William Tang
FTL: Transfer Learning Nonlinear Plasma Dynamic Transitions in Low Dimensional Embeddings via Deep Neural Networks
Apr 26, 2024Abstract:Deep learning algorithms provide a new paradigm to study high-dimensional dynamical behaviors, such as those in fusion plasma systems. Development of novel model reduction methods, coupled with detection of abnormal modes with plasma physics, opens a unique opportunity for building efficient models to identify plasma instabilities for real-time control. Our Fusion Transfer Learning (FTL) model demonstrates success in reconstructing nonlinear kink mode structures by learning from a limited amount of nonlinear simulation data. The knowledge transfer process leverages a pre-trained neural encoder-decoder network, initially trained on linear simulations, to effectively capture nonlinear dynamics. The low-dimensional embeddings extract the coherent structures of interest, while preserving the inherent dynamics of the complex system. Experimental results highlight FTL's capacity to capture transitional behaviors and dynamical features in plasma dynamics -- a task often challenging for conventional methods. The model developed in this study is generalizable and can be extended broadly through transfer learning to address various magnetohydrodynamics (MHD) modes.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
Applications and Techniques for Fast Machine Learning in Science
Oct 25, 2021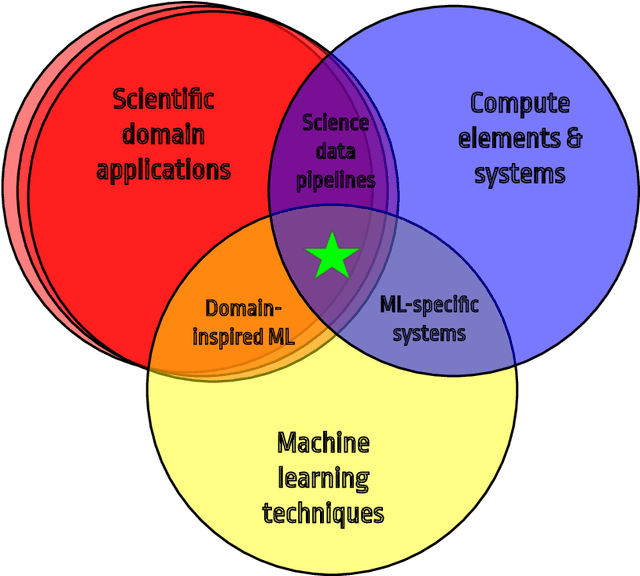
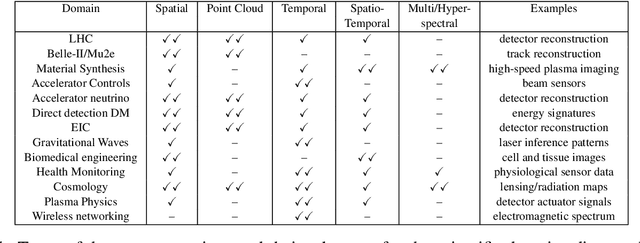
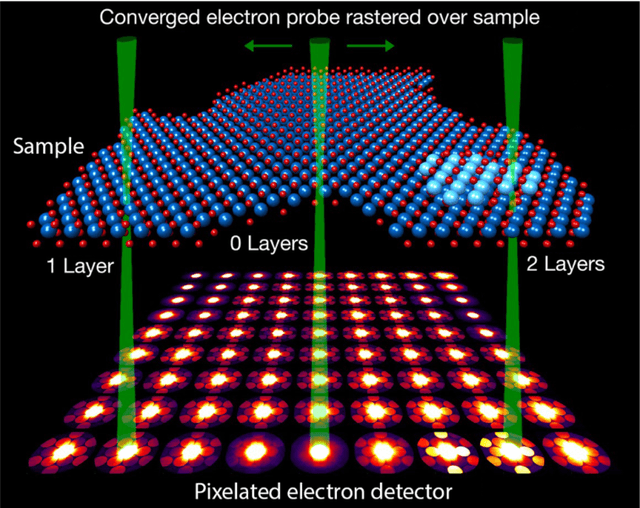
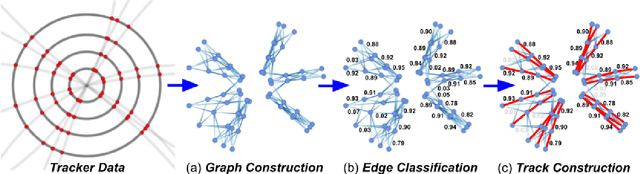
Abstract:In this community review report, we discuss applications and techniques for fast machine learning (ML) in science -- the concept of integrating power ML methods into the real-time experimental data processing loop to accelerate scientific discovery. The material for the report builds on two workshops held by the Fast ML for Science community and covers three main areas: applications for fast ML across a number of scientific domains; techniques for training and implementing performant and resource-efficient ML algorithms; and computing architectures, platforms, and technologies for deploying these algorithms. We also present overlapping challenges across the multiple scientific domains where common solutions can be found. This community report is intended to give plenty of examples and inspiration for scientific discovery through integrated and accelerated ML solutions. This is followed by a high-level overview and organization of technical advances, including an abundance of pointers to source material, which can enable these breakthroughs.
Deep machine learning-assisted multiphoton microscopy to reduce light exposure and expedite imaging
Nov 10, 2020


Abstract:Two-photon excitation fluorescence (2PEF) allows imaging of tissue up to about one millimeter in thickness. Typically, reducing fluorescence excitation exposure reduces the quality of the image. However, using deep learning super resolution techniques, these low-resolution images can be converted to high-resolution images. This work explores improving human tissue imaging by applying deep learning to maximize image quality while reducing fluorescence excitation exposure. We analyze two methods: a method based on U-Net, and a patch-based regression method. Both methods are evaluated on a skin dataset and an eye dataset. The eye dataset includes 1200 paired high power and low power images of retinal organoids. The skin dataset contains multiple frames of each sample of human skin. High-resolution images were formed by averaging 70 frames for each sample and low-resolution images were formed by averaging the first 7 and 15 frames for each sample. The skin dataset includes 550 images for each of the resolution levels. We track two measures of performance for the two methods: mean squared error (MSE) and structural similarity index measure (SSIM). For the eye dataset, the patches method achieves an average MSE of 27,611 compared to 146,855 for the U-Net method, and an average SSIM of 0.636 compared to 0.607 for the U-Net method. For the skin dataset, the patches method achieves an average MSE of 3.768 compared to 4.032 for the U-Net method, and an average SSIM of 0.824 compared to 0.783 for the U-Net method. Despite better performance on image quality, the patches method is worse than the U-Net method when comparing the speed of prediction, taking 303 seconds to predict one image compared to less than one second for the U-Net method.
Training Distributed Deep Recurrent Neural Networks with Mixed Precision on GPU Clusters
Nov 30, 2019



Abstract:In this paper, we evaluate training of deep recurrent neural networks with half-precision floats. We implement a distributed, data-parallel, synchronous training algorithm by integrating TensorFlow and CUDA-aware MPI to enable execution across multiple GPU nodes and making use of high-speed interconnects. We introduce a learning rate schedule facilitating neural network convergence at up to $O(100)$ workers. Strong scaling tests performed on clusters of NVIDIA Pascal P100 GPUs show linear runtime and logarithmic communication time scaling for both single and mixed precision training modes. Performance is evaluated on a scientific dataset taken from the Joint European Torus (JET) tokamak, containing multi-modal time series of sensory measurements leading up to deleterious events called plasma disruptions, and the benchmark Large Movie Review Dataset~\cite{imdb}. Half-precision significantly reduces memory and network bandwidth, allowing training of state-of-the-art models with over 70 million trainable parameters while achieving a comparable test set performance as single precision.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge