Tien-Phat Nguyen
S-Chain: Structured Visual Chain-of-Thought For Medicine
Oct 26, 2025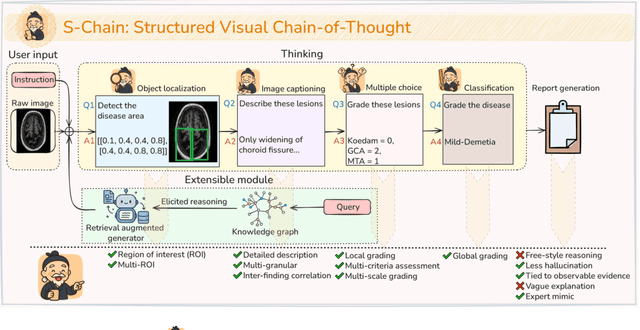
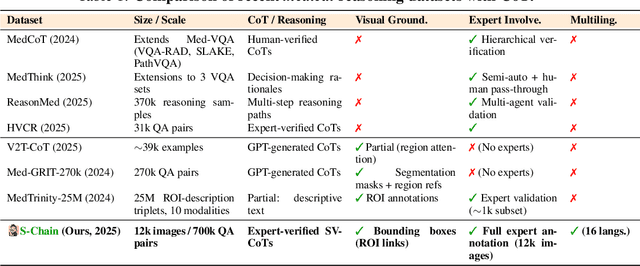
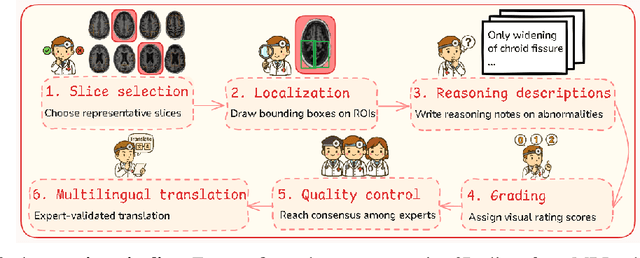
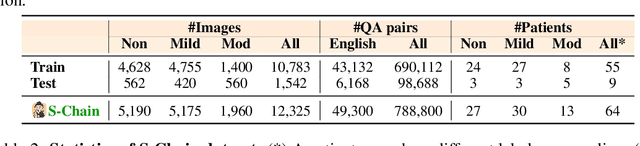
Abstract:Faithful reasoning in medical vision-language models (VLMs) requires not only accurate predictions but also transparent alignment between textual rationales and visual evidence. While Chain-of-Thought (CoT) prompting has shown promise in medical visual question answering (VQA), no large-scale expert-level dataset has captured stepwise reasoning with precise visual grounding. We introduce S-Chain, the first large-scale dataset of 12,000 expert-annotated medical images with bounding boxes and structured visual CoT (SV-CoT), explicitly linking visual regions to reasoning steps. The dataset further supports 16 languages, totaling over 700k VQA pairs for broad multilingual applicability. Using S-Chain, we benchmark state-of-the-art medical VLMs (ExGra-Med, LLaVA-Med) and general-purpose VLMs (Qwen2.5-VL, InternVL2.5), showing that SV-CoT supervision significantly improves interpretability, grounding fidelity, and robustness. Beyond benchmarking, we study its synergy with retrieval-augmented generation, revealing how domain knowledge and visual grounding interact during autoregressive reasoning. Finally, we propose a new mechanism that strengthens the alignment between visual evidence and reasoning, improving both reliability and efficiency. S-Chain establishes a new benchmark for grounded medical reasoning and paves the way toward more trustworthy and explainable medical VLMs.
GazeSearch: Radiology Findings Search Benchmark
Nov 08, 2024



Abstract:Medical eye-tracking data is an important information source for understanding how radiologists visually interpret medical images. This information not only improves the accuracy of deep learning models for X-ray analysis but also their interpretability, enhancing transparency in decision-making. However, the current eye-tracking data is dispersed, unprocessed, and ambiguous, making it difficult to derive meaningful insights. Therefore, there is a need to create a new dataset with more focus and purposeful eyetracking data, improving its utility for diagnostic applications. In this work, we propose a refinement method inspired by the target-present visual search challenge: there is a specific finding and fixations are guided to locate it. After refining the existing eye-tracking datasets, we transform them into a curated visual search dataset, called GazeSearch, specifically for radiology findings, where each fixation sequence is purposefully aligned to the task of locating a particular finding. Subsequently, we introduce a scan path prediction baseline, called ChestSearch, specifically tailored to GazeSearch. Finally, we employ the newly introduced GazeSearch as a benchmark to evaluate the performance of current state-of-the-art methods, offering a comprehensive assessment for visual search in the medical imaging domain.
An objective validation of polyp and instrument segmentation methods in colonoscopy through Medico 2020 polyp segmentation and MedAI 2021 transparency challenges
Jul 30, 2023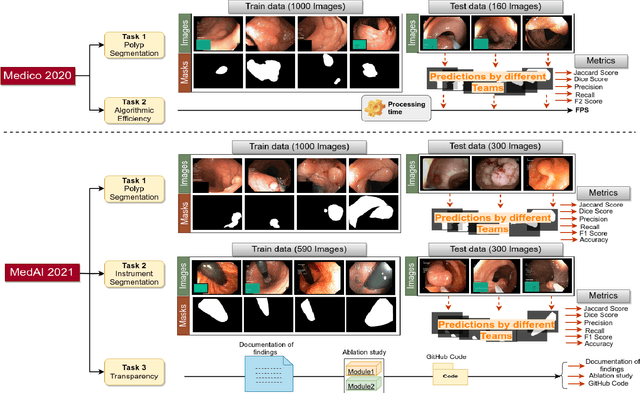
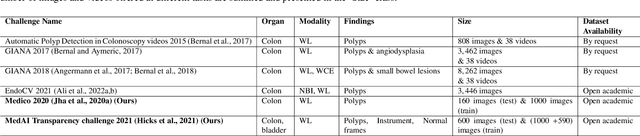
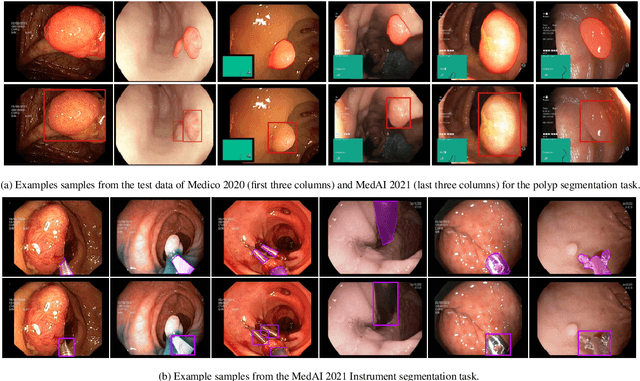
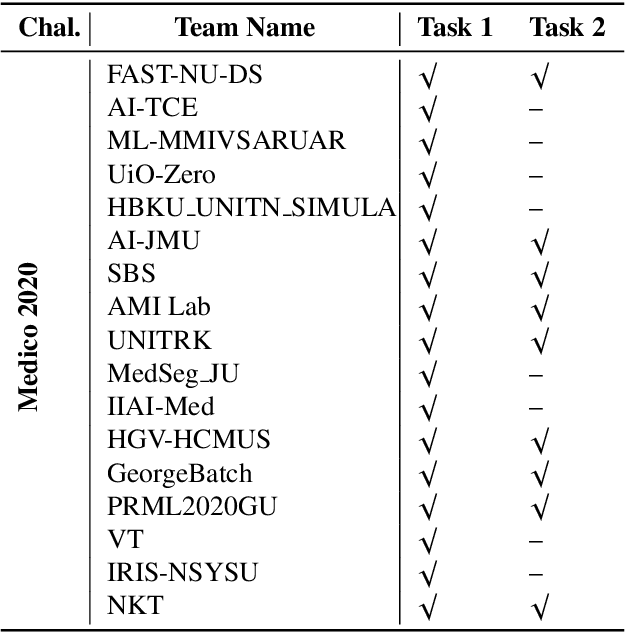
Abstract:Automatic analysis of colonoscopy images has been an active field of research motivated by the importance of early detection of precancerous polyps. However, detecting polyps during the live examination can be challenging due to various factors such as variation of skills and experience among the endoscopists, lack of attentiveness, and fatigue leading to a high polyp miss-rate. Deep learning has emerged as a promising solution to this challenge as it can assist endoscopists in detecting and classifying overlooked polyps and abnormalities in real time. In addition to the algorithm's accuracy, transparency and interpretability are crucial to explaining the whys and hows of the algorithm's prediction. Further, most algorithms are developed in private data, closed source, or proprietary software, and methods lack reproducibility. Therefore, to promote the development of efficient and transparent methods, we have organized the "Medico automatic polyp segmentation (Medico 2020)" and "MedAI: Transparency in Medical Image Segmentation (MedAI 2021)" competitions. We present a comprehensive summary and analyze each contribution, highlight the strength of the best-performing methods, and discuss the possibility of clinical translations of such methods into the clinic. For the transparency task, a multi-disciplinary team, including expert gastroenterologists, accessed each submission and evaluated the team based on open-source practices, failure case analysis, ablation studies, usability and understandability of evaluations to gain a deeper understanding of the models' credibility for clinical deployment. Through the comprehensive analysis of the challenge, we not only highlight the advancements in polyp and surgical instrument segmentation but also encourage qualitative evaluation for building more transparent and understandable AI-based colonoscopy systems.
Z-GMOT: Zero-shot Generic Multiple Object Tracking
May 28, 2023Abstract:Despite the significant progress made in recent years, Multi-Object Tracking (MOT) approaches still suffer from several limitations, including their reliance on prior knowledge of tracking targets, which necessitates the costly annotation of large labeled datasets. As a result, existing MOT methods are limited to a small set of predefined categories, and they struggle with unseen objects in the real world. To address these issues, Generic Multiple Object Tracking (GMOT) has been proposed, which requires less prior information about the targets. However, all existing GMOT approaches follow a one-shot paradigm, relying mainly on the initial bounding box and thus struggling to handle variants e.g., viewpoint, lighting, occlusion, scale, and etc. In this paper, we introduce a novel approach to address the limitations of existing MOT and GMOT methods. Specifically, we propose a zero-shot GMOT (Z-GMOT) algorithm that can track never-seen object categories with zero training examples, without the need for predefined categories or an initial bounding box. To achieve this, we propose iGLIP, an improved version of Grounded language-image pretraining (GLIP), which can detect unseen objects while minimizing false positives. We evaluate our Z-GMOT thoroughly on the GMOT-40 dataset, AnimalTrack testset, DanceTrack testset. The results of these evaluations demonstrate a significant improvement over existing methods. For instance, on the GMOT-40 dataset, the Z-GMOT outperforms one-shot GMOT with OC-SORT by 27.79 points HOTA and 44.37 points MOTA. On the AnimalTrack dataset, it surpasses fully-supervised methods with DeepSORT by 12.55 points HOTA and 8.97 points MOTA. To facilitate further research, we will make our code and models publicly available upon acceptance of this paper.
EmbryosFormer: Deformable Transformer and Collaborative Encoding-Decoding for Embryos Stage Development Classification
Oct 07, 2022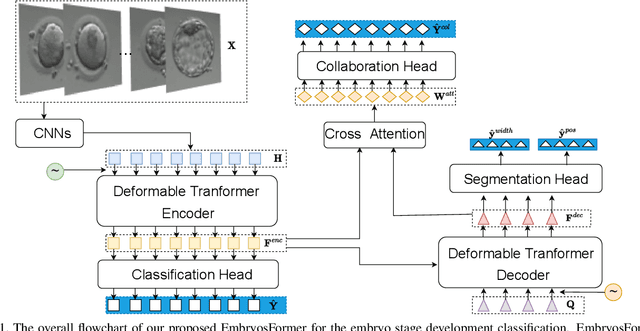
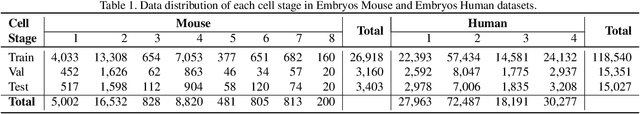
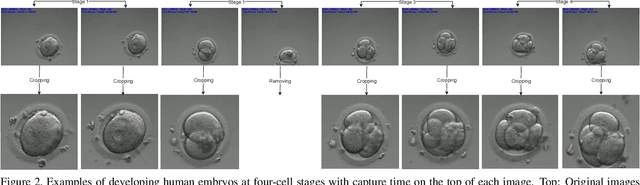
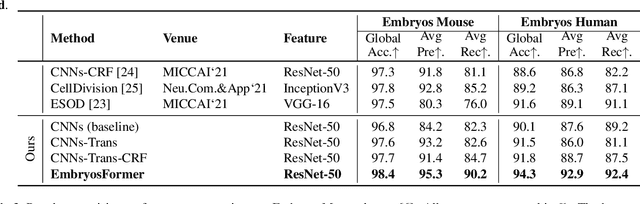
Abstract:The timing of cell divisions in early embryos during the In-Vitro Fertilization (IVF) process is a key predictor of embryo viability. However, observing cell divisions in Time-Lapse Monitoring (TLM) is a time-consuming process and highly depends on experts. In this paper, we propose EmbryosFormer, a computational model to automatically detect and classify cell divisions from original time-lapse images. Our proposed network is designed as an encoder-decoder deformable transformer with collaborative heads. The transformer contracting path predicts per-image labels and is optimized by a classification head. The transformer expanding path models the temporal coherency between embryo images to ensure monotonic non-decreasing constraint and is optimized by a segmentation head. Both contracting and expanding paths are synergetically learned by a collaboration head. We have benchmarked our proposed EmbryosFormer on two datasets: a public dataset with mouse embryos with 8-cell stage and an in-house dataset with human embryos with 4-cell stage. Source code: https://github.com/UARK-AICV/Embryos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge