Sungyoung Lee
GRASP: Guided Region-Aware Sparse Prompting for Adapting MLLMs to Remote Sensing
Jan 23, 2026Abstract:In recent years, Multimodal Large Language Models (MLLMs) have made significant progress in visual question answering tasks. However, directly applying existing fine-tuning methods to remote sensing (RS) images often leads to issues such as overfitting on background noise or neglecting target details. This is primarily due to the large-scale variations, sparse target distributions, and complex regional semantic features inherent in RS images. These challenges limit the effectiveness of MLLMs in RS tasks. To address these challenges, we propose a parameter-efficient fine-tuning (PEFT) strategy called Guided Region-Aware Sparse Prompting (GRASP). GRASP introduces spatially structured soft prompts associated with spatial blocks extracted from a frozen visual token grid. Through a question-guided sparse fusion mechanism, GRASP dynamically aggregates task-specific context into a compact global prompt, enabling the model to focus on relevant regions while filtering out background noise. Extensive experiments on multiple RSVQA benchmarks show that GRASP achieves competitive performance compared to existing fine-tuning and prompt-based methods while maintaining high parameter efficiency.
PPAAS: PVT and Pareto Aware Analog Sizing via Goal-conditioned Reinforcement Learning
Jul 22, 2025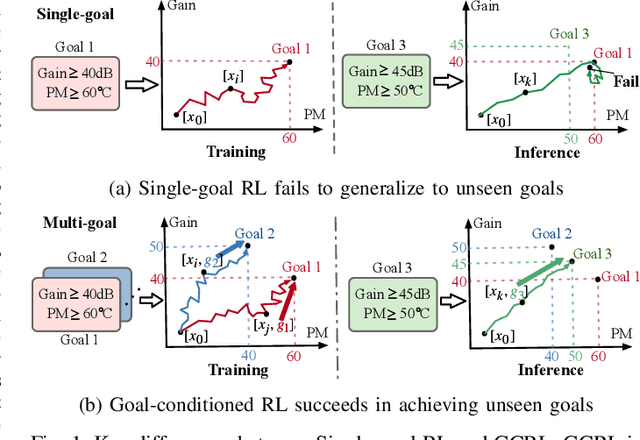
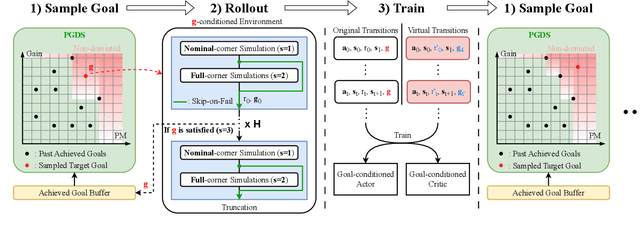
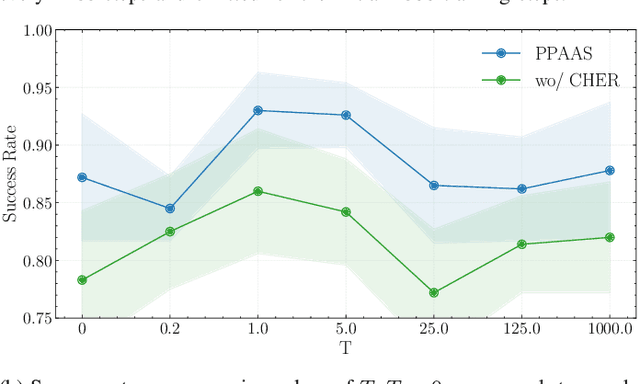
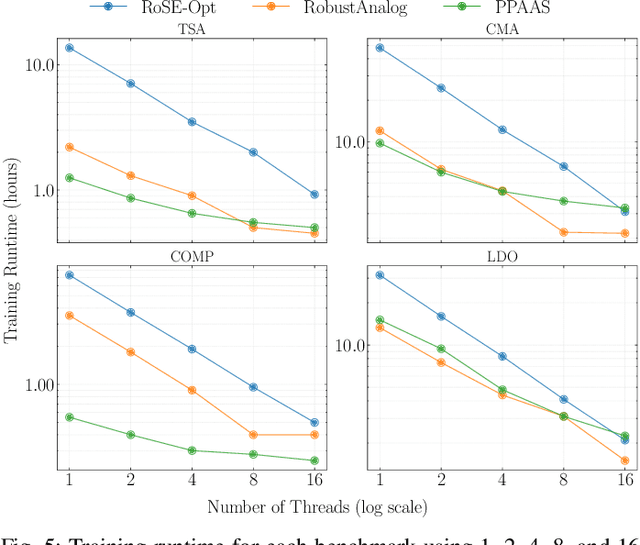
Abstract:Device sizing is a critical yet challenging step in analog and mixed-signal circuit design, requiring careful optimization to meet diverse performance specifications. This challenge is further amplified under process, voltage, and temperature (PVT) variations, which cause circuit behavior to shift across different corners. While reinforcement learning (RL) has shown promise in automating sizing for fixed targets, training a generalized policy that can adapt to a wide range of design specifications under PVT variations requires much more training samples and resources. To address these challenges, we propose a \textbf{Goal-conditioned RL framework} that enables efficient policy training for analog device sizing across PVT corners, with strong generalization capability. To improve sample efficiency, we introduce Pareto-front Dominance Goal Sampling, which constructs an automatic curriculum by sampling goals from the Pareto frontier of previously achieved goals. This strategy is further enhanced by integrating Conservative Hindsight Experience Replay, which assigns relabeled goals with conservative virtual rewards to stabilize training and accelerate convergence. To reduce simulation overhead, our framework incorporates a Skip-on-Fail simulation strategy, which skips full-corner simulations when nominal-corner simulation fails to meet target specifications. Experiments on benchmark circuits demonstrate $\sim$1.6$\times$ improvement in sample efficiency and $\sim$4.1$\times$ improvement in simulation efficiency compared to existing sizing methods. Code and benchmarks are publicly available at https://github.com/SeunggeunKimkr/PPAAS
Self-Supervised Graph Contrastive Pretraining for Device-level Integrated Circuits
Feb 13, 2025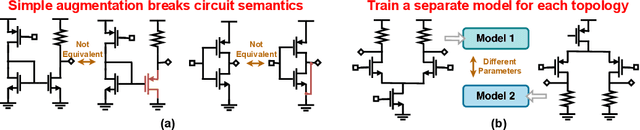
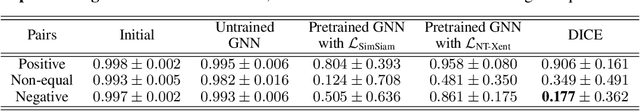
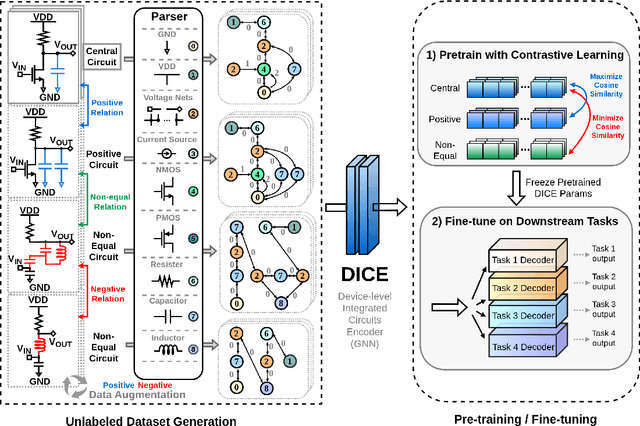
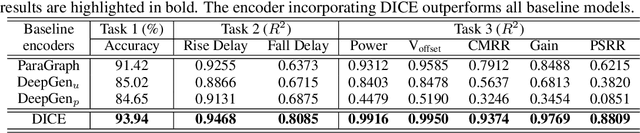
Abstract:Self-supervised graph representation learning has driven significant advancements in domains such as social network analysis, molecular design, and electronics design automation (EDA). However, prior works in EDA have mainly focused on the representation of gate-level digital circuits, failing to capture analog and mixed-signal circuits. To address this gap, we introduce DICE: Device-level Integrated Circuits Encoder, the first self-supervised pretrained graph neural network (GNN) model for any circuit expressed at the device level. DICE is a message-passing neural network (MPNN) trained through graph contrastive learning, and its pretraining process is simulation-free, incorporating two novel data augmentation techniques. Experimental results demonstrate that DICE achieves substantial performance gains across three downstream tasks, underscoring its effectiveness for both analog and digital circuits.
LLaVA Needs More Knowledge: Retrieval Augmented Natural Language Generation with Knowledge Graph for Explaining Thoracic Pathologies
Oct 07, 2024



Abstract:Generating Natural Language Explanations (NLEs) for model predictions on medical images, particularly those depicting thoracic pathologies, remains a critical and challenging task. Existing methodologies often struggle due to general models' insufficient domain-specific medical knowledge and privacy concerns associated with retrieval-based augmentation techniques. To address these issues, we propose a novel Vision-Language framework augmented with a Knowledge Graph (KG)-based datastore, which enhances the model's understanding by incorporating additional domain-specific medical knowledge essential for generating accurate and informative NLEs. Our framework employs a KG-based retrieval mechanism that not only improves the precision of the generated explanations but also preserves data privacy by avoiding direct data retrieval. The KG datastore is designed as a plug-and-play module, allowing for seamless integration with various model architectures. We introduce and evaluate three distinct frameworks within this paradigm: KG-LLaVA, which integrates the pre-trained LLaVA model with KG-RAG; Med-XPT, a custom framework combining MedCLIP, a transformer-based projector, and GPT-2; and Bio-LLaVA, which adapts LLaVA by incorporating the Bio-ViT-L vision model. These frameworks are validated on the MIMIC-NLE dataset, where they achieve state-of-the-art results, underscoring the effectiveness of KG augmentation in generating high-quality NLEs for thoracic pathologies.
AnalogCoder: Analog Circuit Design via Training-Free Code Generation
May 23, 2024



Abstract:Analog circuit design is a significant task in modern chip technology, focusing on the selection of component types, connectivity, and parameters to ensure proper circuit functionality. Despite advances made by Large Language Models (LLMs) in digital circuit design, the complexity and scarcity of data in analog circuitry pose significant challenges. To mitigate these issues, we introduce AnalogCoder, the first training-free LLM agent for designing analog circuits through Python code generation. Firstly, AnalogCoder incorporates a feedback-enhanced flow with tailored domain-specific prompts, enabling the automated and self-correcting design of analog circuits with a high success rate. Secondly, it proposes a circuit tool library to archive successful designs as reusable modular sub-circuits, simplifying composite circuit creation. Thirdly, extensive experiments on a benchmark designed to cover a wide range of analog circuit tasks show that AnalogCoder outperforms other LLM-based methods. It has successfully designed 20 circuits, 5 more than standard GPT-4o. We believe AnalogCoder can significantly improve the labor-intensive chip design process, enabling non-experts to design analog circuits efficiently. Codes and the benchmark are provided at https://github.com/anonyanalog/AnalogCoder.
AI Driven Knowledge Extraction from Clinical Practice Guidelines: Turning Research into Practice
Dec 10, 2020



Abstract:Background and Objectives: Clinical Practice Guidelines (CPGs) represent the foremost methodology for sharing state-of-the-art research findings in the healthcare domain with medical practitioners to limit practice variations, reduce clinical cost, improve the quality of care, and provide evidence based treatment. However, extracting relevant knowledge from the plethora of CPGs is not feasible for already burdened healthcare professionals, leading to large gaps between clinical findings and real practices. It is therefore imperative that state-of-the-art Computing research, especially machine learning is used to provide artificial intelligence based solution for extracting the knowledge from CPGs and reducing the gap between healthcare research/guidelines and practice. Methods: This research presents a novel methodology for knowledge extraction from CPGs to reduce the gap and turn the latest research findings into clinical practice. First, our system classifies the CPG sentences into four classes such as condition-action, condition-consequences, action, and not-applicable based on the information presented in a sentence. We use deep learning with state-of-the-art word embedding, improved word vectors technique in classification process. Second, it identifies qualifier terms in the classified sentences, which assist in recognizing the condition and action phrases in a sentence. Finally, the condition and action phrase are processed and transformed into plain rule If Condition(s) Then Action format. Results: We evaluate the methodology on three different domains guidelines including Hypertension, Rhinosinusitis, and Asthma. The deep learning model classifies the CPG sentences with an accuracy of 95%. While rule extraction was validated by user-centric approach, which achieved a Jaccard coefficient of 0.6, 0.7, and 0.4 with three human experts extracted rules, respectively.
A Practical Approach towards Causality Mining in Clinical Text using Active Transfer Learning
Dec 10, 2020



Abstract:Objective: Causality mining is an active research area, which requires the application of state-of-the-art natural language processing techniques. In the healthcare domain, medical experts create clinical text to overcome the limitation of well-defined and schema driven information systems. The objective of this research work is to create a framework, which can convert clinical text into causal knowledge. Methods: A practical approach based on term expansion, phrase generation, BERT based phrase embedding and semantic matching, semantic enrichment, expert verification, and model evolution has been used to construct a comprehensive causality mining framework. This active transfer learning based framework along with its supplementary services, is able to extract and enrich, causal relationships and their corresponding entities from clinical text. Results: The multi-model transfer learning technique when applied over multiple iterations, gains performance improvements in terms of its accuracy and recall while keeping the precision constant. We also present a comparative analysis of the presented techniques with their common alternatives, which demonstrate the correctness of our approach and its ability to capture most causal relationships. Conclusion: The presented framework has provided cutting-edge results in the healthcare domain. However, the framework can be tweaked to provide causality detection in other domains, as well. Significance: The presented framework is generic enough to be utilized in any domain, healthcare services can gain massive benefits due to the voluminous and various nature of its data. This causal knowledge extraction framework can be used to summarize clinical text, create personas, discover medical knowledge, and provide evidence to clinical decision making.
Precision Medicine Informatics: Principles, Prospects, and Challenges
Nov 04, 2019



Abstract:Precision Medicine (PM) is an emerging approach that appears with the impression of changing the existing paradigm of medical practice. Recent advances in technological innovations and genetics, and the growing availability of health data have set a new pace of the research and imposes a set of new requirements on different stakeholders. To date, some studies are available that discuss about different aspects of PM. Nevertheless, a holistic representation of those aspects deemed to confer the technological perspective, in relation to applications and challenges, is mostly ignored. In this context, this paper surveys advances in PM from informatics viewpoint and reviews the enabling tools and techniques in a categorized manner. In addition, the study discusses how other technological paradigms including big data, artificial intelligence, and internet of things can be exploited to advance the potentials of PM. Furthermore, the paper provides some guidelines for future research for seamless implementation and wide-scale deployment of PM based on identified open issues and associated challenges. To this end, the paper proposes an integrated holistic framework for PM motivating informatics researchers to design their relevant research works in an appropriate context.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge