Yong Hyun Ahn
LLaVA Needs More Knowledge: Retrieval Augmented Natural Language Generation with Knowledge Graph for Explaining Thoracic Pathologies
Oct 07, 2024



Abstract:Generating Natural Language Explanations (NLEs) for model predictions on medical images, particularly those depicting thoracic pathologies, remains a critical and challenging task. Existing methodologies often struggle due to general models' insufficient domain-specific medical knowledge and privacy concerns associated with retrieval-based augmentation techniques. To address these issues, we propose a novel Vision-Language framework augmented with a Knowledge Graph (KG)-based datastore, which enhances the model's understanding by incorporating additional domain-specific medical knowledge essential for generating accurate and informative NLEs. Our framework employs a KG-based retrieval mechanism that not only improves the precision of the generated explanations but also preserves data privacy by avoiding direct data retrieval. The KG datastore is designed as a plug-and-play module, allowing for seamless integration with various model architectures. We introduce and evaluate three distinct frameworks within this paradigm: KG-LLaVA, which integrates the pre-trained LLaVA model with KG-RAG; Med-XPT, a custom framework combining MedCLIP, a transformer-based projector, and GPT-2; and Bio-LLaVA, which adapts LLaVA by incorporating the Bio-ViT-L vision model. These frameworks are validated on the MIMIC-NLE dataset, where they achieve state-of-the-art results, underscoring the effectiveness of KG augmentation in generating high-quality NLEs for thoracic pathologies.
Mask-Free Neuron Concept Annotation for Interpreting Neural Networks in Medical Domain
Jul 16, 2024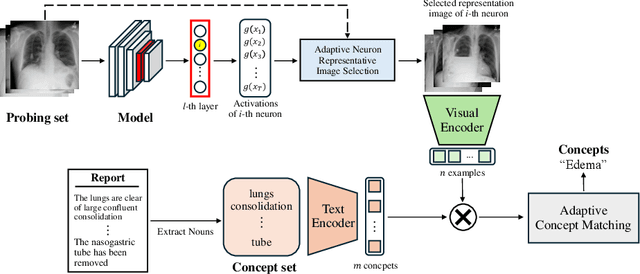

Abstract:Recent advancements in deep neural networks have shown promise in aiding disease diagnosis and medical decision-making. However, ensuring transparent decision-making processes of AI models in compliance with regulations requires a comprehensive understanding of the model's internal workings. However, previous methods heavily rely on expensive pixel-wise annotated datasets for interpreting the model, presenting a significant drawback in medical domains. In this paper, we propose a novel medical neuron concept annotation method, named Mask-free Medical Model Interpretation (MAMMI), addresses these challenges. By using a vision-language model, our method relaxes the need for pixel-level masks for neuron concept annotation. MAMMI achieves superior performance compared to other interpretation methods, demonstrating its efficacy in providing rich representations for neurons in medical image analysis. Our experiments on a model trained on NIH chest X-rays validate the effectiveness of MAMMI, showcasing its potential for transparent clinical decision-making in the medical domain. The code is available at https://github.com/ailab-kyunghee/MAMMI.
WWW: A Unified Framework for Explaining What, Where and Why of Neural Networks by Interpretation of Neuron Concepts
Feb 29, 2024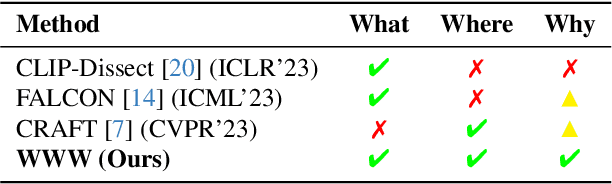
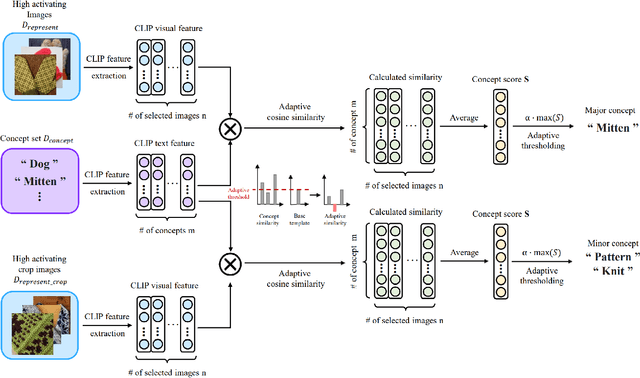
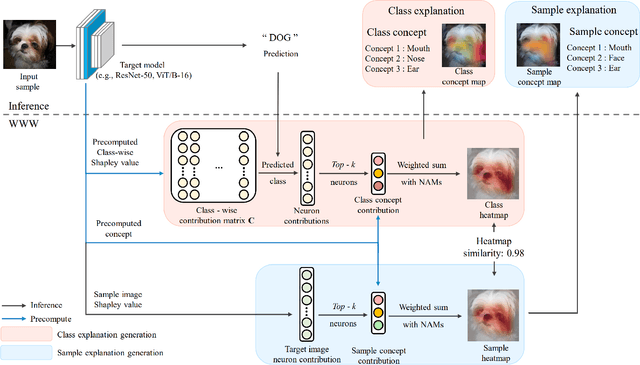
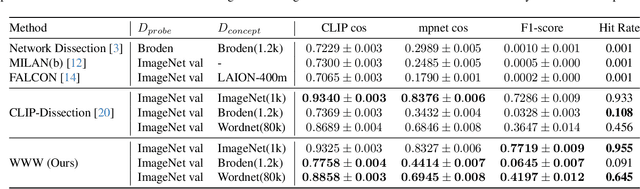
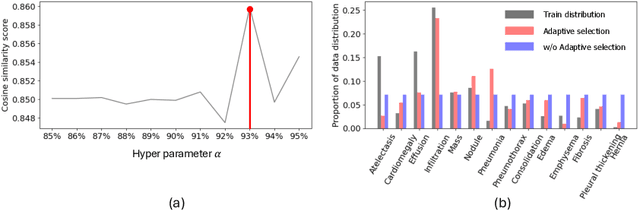
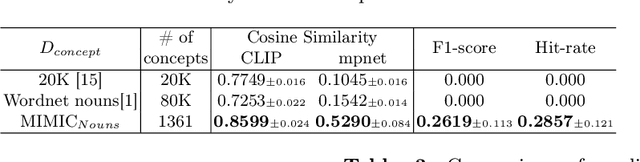
Abstract:Recent advancements in neural networks have showcased their remarkable capabilities across various domains. Despite these successes, the "black box" problem still remains. Addressing this, we propose a novel framework, WWW, that offers the 'what', 'where', and 'why' of the neural network decisions in human-understandable terms. Specifically, WWW utilizes adaptive selection for concept discovery, employing adaptive cosine similarity and thresholding techniques to effectively explain 'what'. To address the 'where' and 'why', we proposed a novel combination of neuron activation maps (NAMs) with Shapley values, generating localized concept maps and heatmaps for individual inputs. Furthermore, WWW introduces a method for predicting uncertainty, leveraging heatmap similarities to estimate 'how' reliable the prediction is. Experimental evaluations of WWW demonstrate superior performance in both quantitative and qualitative metrics, outperforming existing methods in interpretability. WWW provides a unified solution for explaining 'what', 'where', and 'why', introducing a method for localized explanations from global interpretations and offering a plug-and-play solution adaptable to various architectures.
LINe: Out-of-Distribution Detection by Leveraging Important Neurons
Mar 24, 2023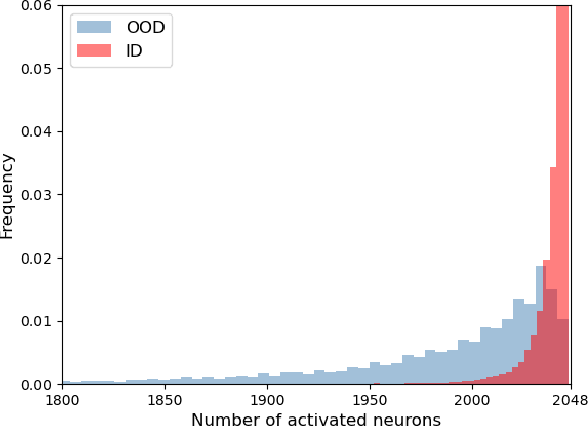
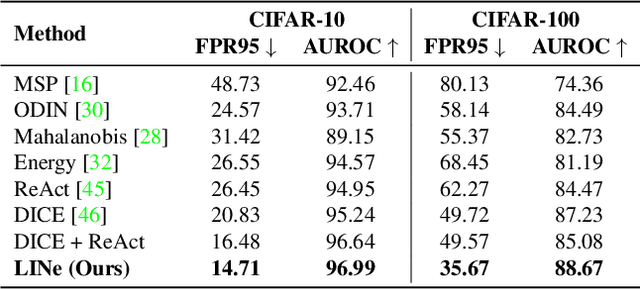
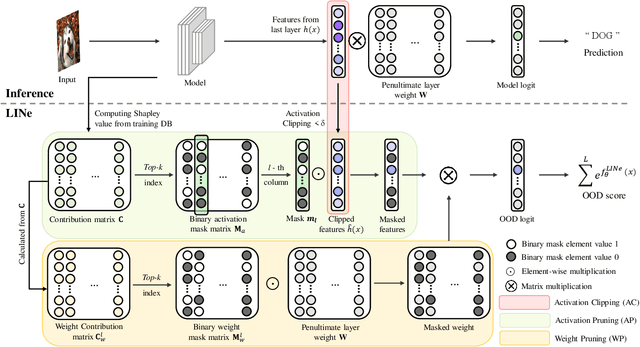
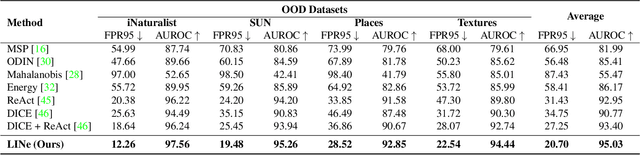
Abstract:It is important to quantify the uncertainty of input samples, especially in mission-critical domains such as autonomous driving and healthcare, where failure predictions on out-of-distribution (OOD) data are likely to cause big problems. OOD detection problem fundamentally begins in that the model cannot express what it is not aware of. Post-hoc OOD detection approaches are widely explored because they do not require an additional re-training process which might degrade the model's performance and increase the training cost. In this study, from the perspective of neurons in the deep layer of the model representing high-level features, we introduce a new aspect for analyzing the difference in model outputs between in-distribution data and OOD data. We propose a novel method, Leveraging Important Neurons (LINe), for post-hoc Out of distribution detection. Shapley value-based pruning reduces the effects of noisy outputs by selecting only high-contribution neurons for predicting specific classes of input data and masking the rest. Activation clipping fixes all values above a certain threshold into the same value, allowing LINe to treat all the class-specific features equally and just consider the difference between the number of activated feature differences between in-distribution and OOD data. Comprehensive experiments verify the effectiveness of the proposed method by outperforming state-of-the-art post-hoc OOD detection methods on CIFAR-10, CIFAR-100, and ImageNet datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge