Ameer Hamza
Autonomous AI Surveillance: Multimodal Deep Learning for Cognitive and Behavioral Monitoring
Jul 02, 2025
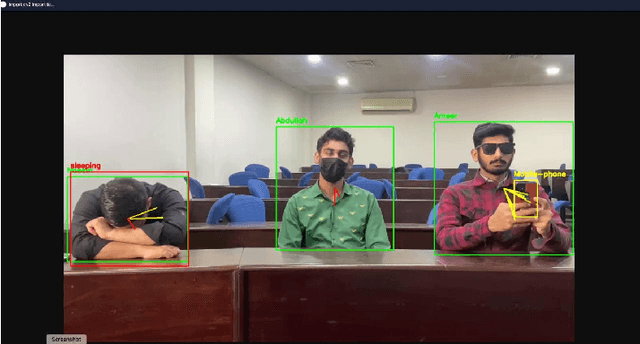
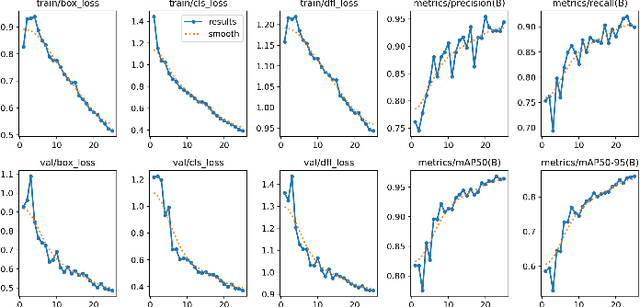
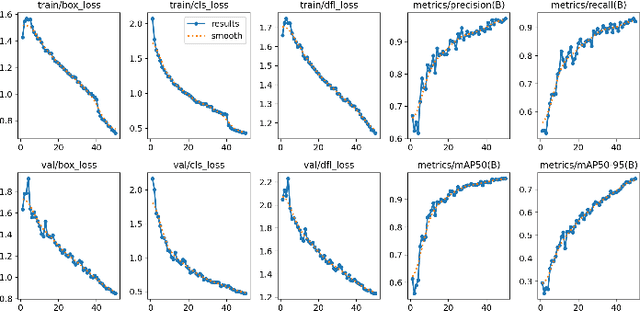
Abstract:This study presents a novel classroom surveillance system that integrates multiple modalities, including drowsiness, tracking of mobile phone usage, and face recognition,to assess student attentiveness with enhanced precision.The system leverages the YOLOv8 model to detect both mobile phone and sleep usage,(Ghatge et al., 2024) while facial recognition is achieved through LResNet Occ FC body tracking using YOLO and MTCNN.(Durai et al., 2024) These models work in synergy to provide comprehensive, real-time monitoring, offering insights into student engagement and behavior.(S et al., 2023) The framework is trained on specialized datasets, such as the RMFD dataset for face recognition and a Roboflow dataset for mobile phone detection. The extensive evaluation of the system shows promising results. Sleep detection achieves 97. 42% mAP@50, face recognition achieves 86. 45% validation accuracy and mobile phone detection reach 85. 89% mAP@50. The system is implemented within a core PHP web application and utilizes ESP32-CAM hardware for seamless data capture.(Neto et al., 2024) This integrated approach not only enhances classroom monitoring, but also ensures automatic attendance recording via face recognition as students remain seated in the classroom, offering scalability for diverse educational environments.(Banada,2025)
Natural Language Processing for Analyzing Electronic Health Records and Clinical Notes in Cancer Research: A Review
Oct 29, 2024Abstract:Objective: This review aims to analyze the application of natural language processing (NLP) techniques in cancer research using electronic health records (EHRs) and clinical notes. This review addresses gaps in the existing literature by providing a broader perspective than previous studies focused on specific cancer types or applications. Methods: A comprehensive literature search was conducted using the Scopus database, identifying 94 relevant studies published between 2019 and 2024. Data extraction included study characteristics, cancer types, NLP methodologies, dataset information, performance metrics, challenges, and future directions. Studies were categorized based on cancer types and NLP applications. Results: The results showed a growing trend in NLP applications for cancer research, with breast, lung, and colorectal cancers being the most studied. Information extraction and text classification emerged as predominant NLP tasks. A shift from rule-based to advanced machine learning techniques, particularly transformer-based models, was observed. The Dataset sizes used in existing studies varied widely. Key challenges included the limited generalizability of proposed solutions and the need for improved integration into clinical workflows. Conclusion: NLP techniques show significant potential in analyzing EHRs and clinical notes for cancer research. However, future work should focus on improving model generalizability, enhancing robustness in handling complex clinical language, and expanding applications to understudied cancer types. Integration of NLP tools into clinical practice and addressing ethical considerations remain crucial for utilizing the full potential of NLP in enhancing cancer diagnosis, treatment, and patient outcomes.
Resource-Efficient Medical Report Generation using Large Language Models
Oct 21, 2024Abstract:Medical report generation is the task of automatically writing radiology reports for chest X-ray images. Manually composing these reports is a time-consuming process that is also prone to human errors. Generating medical reports can therefore help reduce the burden on radiologists. In other words, we can promote greater clinical automation in the medical domain. In this work, we propose a new framework leveraging vision-enabled Large Language Models (LLM) for the task of medical report generation. We introduce a lightweight solution that achieves better or comparative performance as compared to previous solutions on the task of medical report generation. We conduct extensive experiments exploring different model sizes and enhancement approaches, such as prefix tuning to improve the text generation abilities of the LLMs. We evaluate our approach on a prominent large-scale radiology report dataset - MIMIC-CXR. Our results demonstrate the capability of our resource-efficient framework to generate patient-specific reports with strong medical contextual understanding and high precision.
LLaVA Needs More Knowledge: Retrieval Augmented Natural Language Generation with Knowledge Graph for Explaining Thoracic Pathologies
Oct 07, 2024



Abstract:Generating Natural Language Explanations (NLEs) for model predictions on medical images, particularly those depicting thoracic pathologies, remains a critical and challenging task. Existing methodologies often struggle due to general models' insufficient domain-specific medical knowledge and privacy concerns associated with retrieval-based augmentation techniques. To address these issues, we propose a novel Vision-Language framework augmented with a Knowledge Graph (KG)-based datastore, which enhances the model's understanding by incorporating additional domain-specific medical knowledge essential for generating accurate and informative NLEs. Our framework employs a KG-based retrieval mechanism that not only improves the precision of the generated explanations but also preserves data privacy by avoiding direct data retrieval. The KG datastore is designed as a plug-and-play module, allowing for seamless integration with various model architectures. We introduce and evaluate three distinct frameworks within this paradigm: KG-LLaVA, which integrates the pre-trained LLaVA model with KG-RAG; Med-XPT, a custom framework combining MedCLIP, a transformer-based projector, and GPT-2; and Bio-LLaVA, which adapts LLaVA by incorporating the Bio-ViT-L vision model. These frameworks are validated on the MIMIC-NLE dataset, where they achieve state-of-the-art results, underscoring the effectiveness of KG augmentation in generating high-quality NLEs for thoracic pathologies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge