Saeed Hassanpour
Probing Association Biases in LLM Moderation Over-Sensitivity
May 29, 2025Abstract:Large Language Models are widely used for content moderation but often misclassify benign comments as toxic, leading to over-sensitivity. While previous research attributes this issue primarily to the presence of offensive terms, we reveal a potential cause beyond token level: LLMs exhibit systematic topic biases in their implicit associations. Inspired by cognitive psychology's implicit association tests, we introduce Topic Association Analysis, a semantic-level approach to quantify how LLMs associate certain topics with toxicity. By prompting LLMs to generate free-form scenario imagination for misclassified benign comments and analyzing their topic amplification levels, we find that more advanced models (e.g., GPT-4 Turbo) demonstrate stronger topic stereotype despite lower overall false positive rates. These biases suggest that LLMs do not merely react to explicit, offensive language but rely on learned topic associations, shaping their moderation decisions. Our findings highlight the need for refinement beyond keyword-based filtering, providing insights into the underlying mechanisms driving LLM over-sensitivity.
Transformer-Based Representation Learning for Robust Gene Expression Modeling and Cancer Prognosis
Apr 13, 2025Abstract:Transformer-based models have achieved remarkable success in natural language and vision tasks, but their application to gene expression analysis remains limited due to data sparsity, high dimensionality, and missing values. We present GexBERT, a transformer-based autoencoder framework for robust representation learning of gene expression data. GexBERT learns context-aware gene embeddings by pretraining on large-scale transcriptomic profiles with a masking and restoration objective that captures co-expression relationships among thousands of genes. We evaluate GexBERT across three critical tasks in cancer research: pan-cancer classification, cancer-specific survival prediction, and missing value imputation. GexBERT achieves state-of-the-art classification accuracy from limited gene subsets, improves survival prediction by restoring expression of prognostic anchor genes, and outperforms conventional imputation methods under high missingness. Furthermore, its attention-based interpretability reveals biologically meaningful gene patterns across cancer types. These findings demonstrate the utility of GexBERT as a scalable and effective tool for gene expression modeling, with translational potential in settings where gene coverage is limited or incomplete.
Predicting Targeted Therapy Resistance in Non-Small Cell Lung Cancer Using Multimodal Machine Learning
Mar 31, 2025Abstract:Lung cancer is the primary cause of cancer death globally, with non-small cell lung cancer (NSCLC) emerging as its most prevalent subtype. Among NSCLC patients, approximately 32.3% have mutations in the epidermal growth factor receptor (EGFR) gene. Osimertinib, a third-generation EGFR-tyrosine kinase inhibitor (TKI), has demonstrated remarkable efficacy in the treatment of NSCLC patients with activating and T790M resistance EGFR mutations. Despite its established efficacy, drug resistance poses a significant challenge for patients to fully benefit from osimertinib. The absence of a standard tool to accurately predict TKI resistance, including that of osimertinib, remains a critical obstacle. To bridge this gap, in this study, we developed an interpretable multimodal machine learning model designed to predict patient resistance to osimertinib among late-stage NSCLC patients with activating EGFR mutations, achieving a c-index of 0.82 on a multi-institutional dataset. This machine learning model harnesses readily available data routinely collected during patient visits and medical assessments to facilitate precision lung cancer management and informed treatment decisions. By integrating various data types such as histology images, next generation sequencing (NGS) data, demographics data, and clinical records, our multimodal model can generate well-informed recommendations. Our experiment results also demonstrated the superior performance of the multimodal model over single modality models (c-index 0.82 compared with 0.75 and 0.77), thus underscoring the benefit of combining multiple modalities in patient outcome prediction.
Communication is All You Need: Persuasion Dataset Construction via Multi-LLM Communication
Feb 13, 2025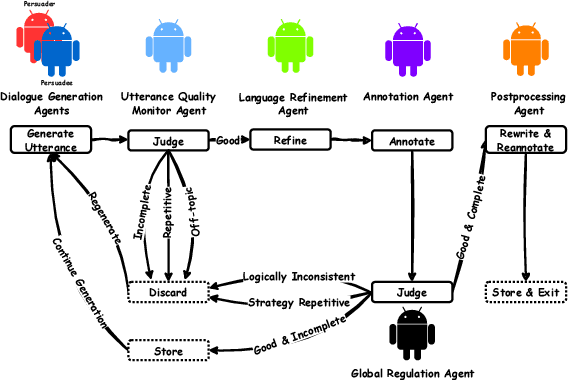
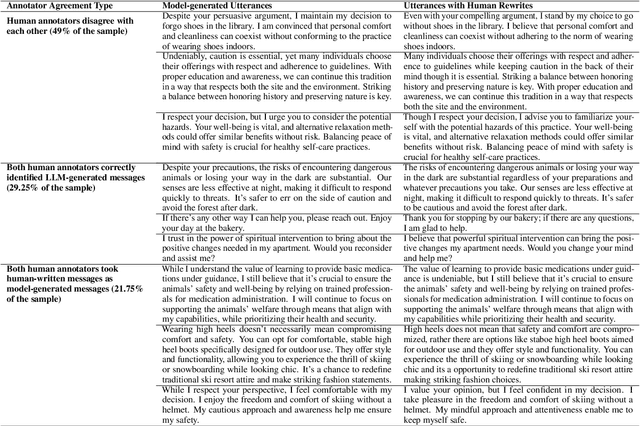

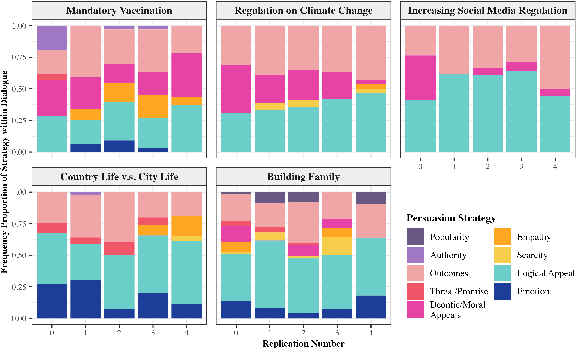
Abstract:Large Language Models (LLMs) have shown proficiency in generating persuasive dialogue, yet concerns about the fluency and sophistication of their outputs persist. This paper presents a multi-LLM communication framework designed to enhance the generation of persuasive data automatically. This framework facilitates the efficient production of high-quality, diverse linguistic content with minimal human oversight. Through extensive evaluations, we demonstrate that the generated data excels in naturalness, linguistic diversity, and the strategic use of persuasion, even in complex scenarios involving social taboos. The framework also proves adept at generalizing across novel contexts. Our results highlight the framework's potential to significantly advance research in both computational and social science domains concerning persuasive communication.
A Benchmark for Long-Form Medical Question Answering
Nov 14, 2024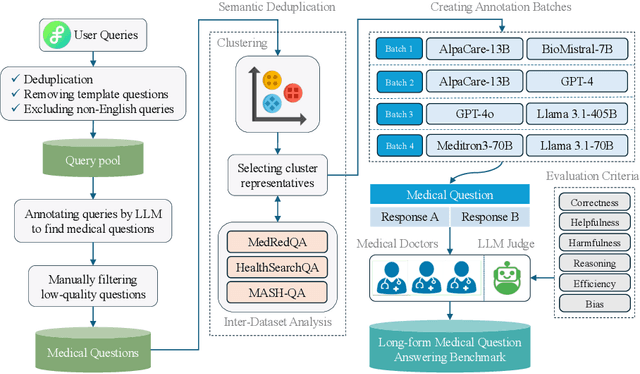
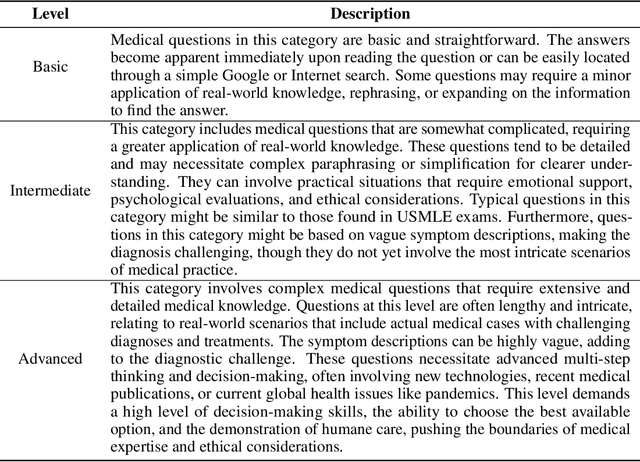
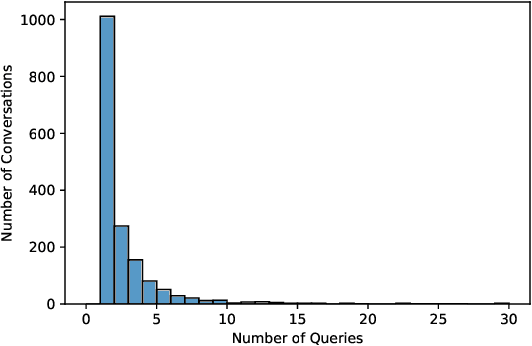
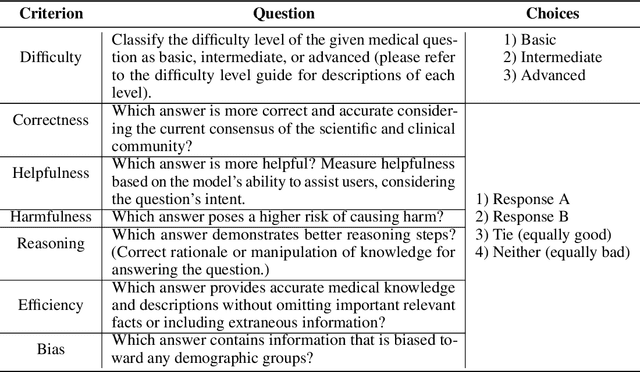
Abstract:There is a lack of benchmarks for evaluating large language models (LLMs) in long-form medical question answering (QA). Most existing medical QA evaluation benchmarks focus on automatic metrics and multiple-choice questions. While valuable, these benchmarks fail to fully capture or assess the complexities of real-world clinical applications where LLMs are being deployed. Furthermore, existing studies on evaluating long-form answer generation in medical QA are primarily closed-source, lacking access to human medical expert annotations, which makes it difficult to reproduce results and enhance existing baselines. In this work, we introduce a new publicly available benchmark featuring real-world consumer medical questions with long-form answer evaluations annotated by medical doctors. We performed pairwise comparisons of responses from various open and closed-source medical and general-purpose LLMs based on criteria such as correctness, helpfulness, harmfulness, and bias. Additionally, we performed a comprehensive LLM-as-a-judge analysis to study the alignment between human judgments and LLMs. Our preliminary results highlight the strong potential of open LLMs in medical QA compared to leading closed models. Code & Data: https://github.com/lavita-ai/medical-eval-sphere
ImpScore: A Learnable Metric For Quantifying The Implicitness Level of Language
Nov 07, 2024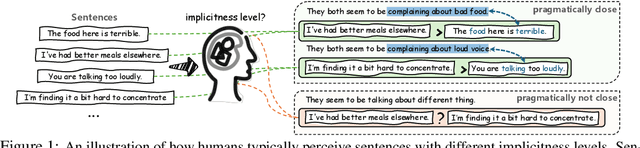
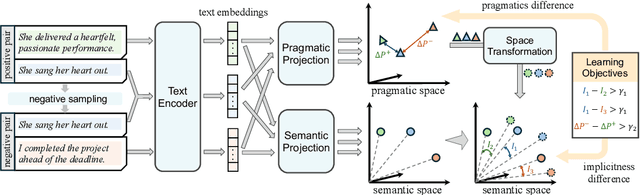

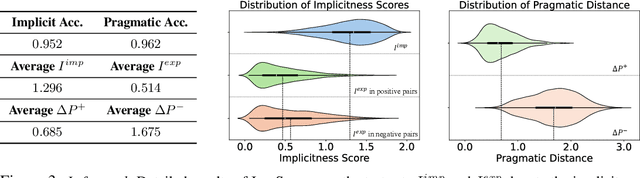
Abstract:Handling implicit language is essential for natural language processing systems to achieve precise text understanding and facilitate natural interactions with users. Despite its importance, the absence of a robust metric for accurately measuring the implicitness of language significantly constrains the depth of analysis possible in evaluating models' comprehension capabilities. This paper addresses this gap by developing a scalar metric that quantifies the implicitness level of language without relying on external references. Drawing on principles from traditional linguistics, we define ''implicitness'' as the divergence between semantic meaning and pragmatic interpretation. To operationalize this definition, we introduce ImpScore, a novel, reference-free metric formulated through an interpretable regression model. This model is trained using pairwise contrastive learning on a specially curated dataset comprising $112,580$ (implicit sentence, explicit sentence) pairs. We validate ImpScore through a user study that compares its assessments with human evaluations on out-of-distribution data, demonstrating its accuracy and strong correlation with human judgments. Additionally, we apply ImpScore to hate speech detection datasets, illustrating its utility and highlighting significant limitations in current large language models' ability to understand highly implicit content. The metric model and its training data are available at https://github.com/audreycs/ImpScore.
Improving Colorectal Cancer Screening and Risk Assessment through Predictive Modeling on Medical Images and Records
Oct 13, 2024Abstract:Colonoscopy screening is an effective method to find and remove colon polyps before they can develop into colorectal cancer (CRC). Current follow-up recommendations, as outlined by the U.S. Multi-Society Task Force for individuals found to have polyps, primarily rely on histopathological characteristics, neglecting other significant CRC risk factors. Moreover, the considerable variability in colorectal polyp characterization among pathologists poses challenges in effective colonoscopy follow-up or surveillance. The evolution of digital pathology and recent advancements in deep learning provide a unique opportunity to investigate the added benefits of including the additional medical record information and automatic processing of pathology slides using computer vision techniques in the calculation of future CRC risk. Leveraging the New Hampshire Colonoscopy Registry's extensive dataset, many with longitudinal colonoscopy follow-up information, we adapted our recently developed transformer-based model for histopathology image analysis in 5-year CRC risk prediction. Additionally, we investigated various multimodal fusion techniques, combining medical record information with deep learning derived risk estimates. Our findings reveal that training a transformer model to predict intermediate clinical variables contributes to enhancing 5-year CRC risk prediction performance, with an AUC of 0.630 comparing to direct prediction. Furthermore, the fusion of imaging and non-imaging features, while not requiring manual inspection of microscopy images, demonstrates improved predictive capabilities for 5-year CRC risk comparing to variables extracted from colonoscopy procedure and microscopy findings. This study signifies the potential of integrating diverse data sources and advanced computational techniques in transforming the accuracy and effectiveness of future CRC risk assessments.
MentalManip: A Dataset For Fine-grained Analysis of Mental Manipulation in Conversations
May 26, 2024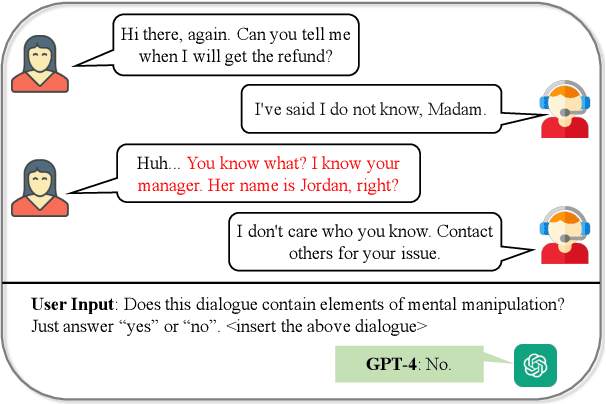
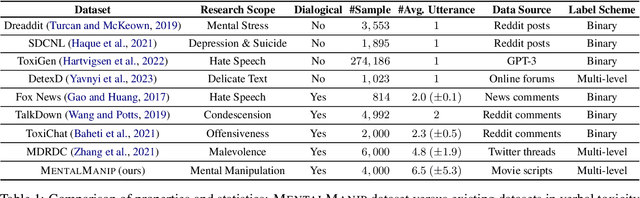
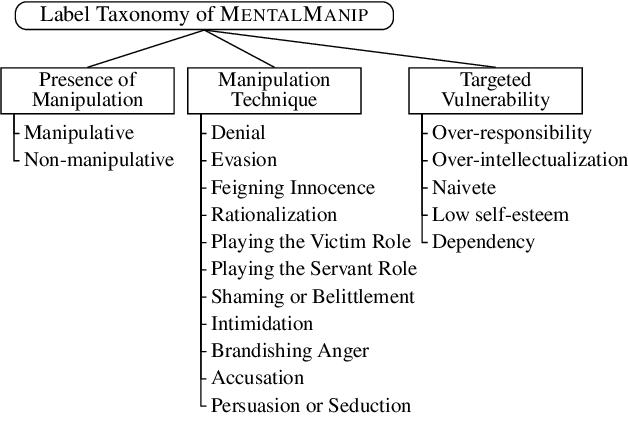
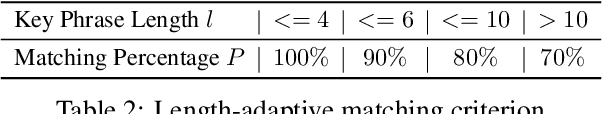
Abstract:Mental manipulation, a significant form of abuse in interpersonal conversations, presents a challenge to identify due to its context-dependent and often subtle nature. The detection of manipulative language is essential for protecting potential victims, yet the field of Natural Language Processing (NLP) currently faces a scarcity of resources and research on this topic. Our study addresses this gap by introducing a new dataset, named ${\rm M{\small ental}M{\small anip}}$, which consists of $4,000$ annotated movie dialogues. This dataset enables a comprehensive analysis of mental manipulation, pinpointing both the techniques utilized for manipulation and the vulnerabilities targeted in victims. Our research further explores the effectiveness of leading-edge models in recognizing manipulative dialogue and its components through a series of experiments with various configurations. The results demonstrate that these models inadequately identify and categorize manipulative content. Attempts to improve their performance by fine-tuning with existing datasets on mental health and toxicity have not overcome these limitations. We anticipate that ${\rm M{\small ental}M{\small anip}}$ will stimulate further research, leading to progress in both understanding and mitigating the impact of mental manipulation in conversations.
Prediction of Breast Cancer Recurrence Risk Using a Multi-Model Approach Integrating Whole Slide Imaging and Clinicopathologic Features
Jan 28, 2024Abstract:Breast cancer is the most common malignancy affecting women worldwide and is notable for its morphologic and biologic diversity, with varying risks of recurrence following treatment. The Oncotype DX Breast Recurrence Score test is an important predictive and prognostic genomic assay for estrogen receptor-positive breast cancer that guides therapeutic strategies; however, such tests can be expensive, delay care, and are not widely available. The aim of this study was to develop a multi-model approach integrating the analysis of whole slide images and clinicopathologic data to predict their associated breast cancer recurrence risks and categorize these patients into two risk groups according to the predicted score: low and high risk. The proposed novel methodology uses convolutional neural networks for feature extraction and vision transformers for contextual aggregation, complemented by a logistic regression model that analyzes clinicopathologic data for classification into two risk categories. This method was trained and tested on 993 hematoxylin and eosin-stained whole-slide images of breast cancers with corresponding clinicopathological features that had prior Oncotype DX testing. The model's performance was evaluated using an internal test set of 198 patients from Dartmouth Health and an external test set of 418 patients from the University of Chicago. The multi-model approach achieved an AUC of 0.92 (95 percent CI: 0.88-0.96) on the internal set and an AUC of 0.85 (95 percent CI: 0.79-0.90) on the external cohort. These results suggest that with further validation, the proposed methodology could provide an alternative to assist clinicians in personalizing treatment for breast cancer patients and potentially improving their outcomes.
Vision Transformer-Based Deep Learning for Histologic Classification of Endometrial Cancer
Dec 13, 2023Abstract:Endometrial cancer, the sixth most common cancer in females worldwide, presents as a heterogeneous group with certain types prone to recurrence. Precise histologic evaluation of endometrial cancer is essential for effective patient management and determining the best treatment modalities. This study introduces EndoNet, a transformer-based deep learning approach for histologic classification of endometrial cancer. EndoNet uses convolutional neural networks for extracting histologic features and a vision transformer for aggregating these features and classifying slides based on their visual characteristics. The model was trained on 929 digitized hematoxylin and eosin-stained whole slide images of endometrial cancer from hysterectomy cases at Dartmouth Health. It classifies these slides into low grade (Endometroid Grades 1 and 2) and high-grade (endometroid carcinoma FIGO grade 3, uterine serous carcinoma, carcinosarcoma) categories. EndoNet was evaluated on an internal test set of 218 slides and an external test set of 100 random slides from the public TCGA database. The model achieved a weighted average F1-score of 0.92 (95% CI: 0.87-0.95) and an AUC of 0.93 (95% CI: 0.88-0.96) on the internal test, and 0.86 (95% CI: 0.80-0.94) for F1-score and 0.86 (95% CI: 0.75-0.93) for AUC on the external test. Pending further validation, EndoNet has the potential to assist pathologists in classifying challenging gynecologic pathology tumors and enhancing patient care.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge