Bing Ren
A Benchmark for Long-Form Medical Question Answering
Nov 14, 2024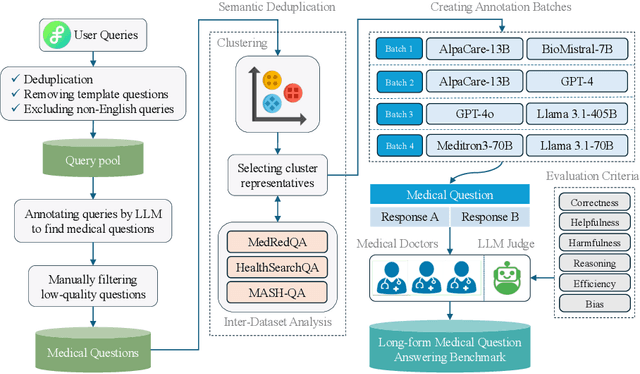
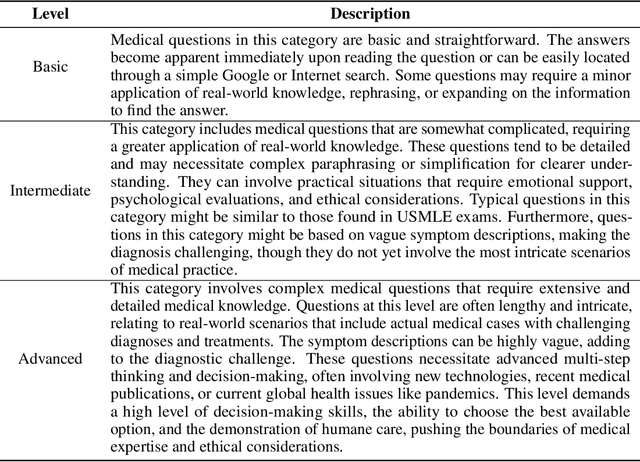
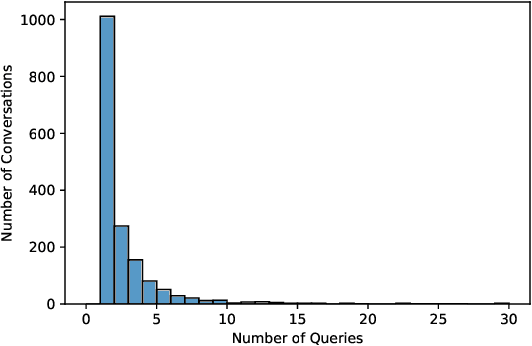
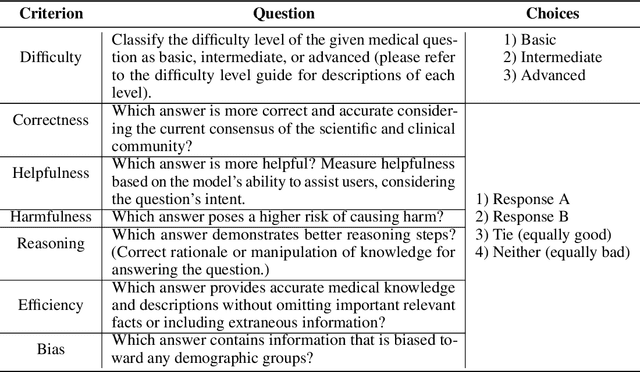
Abstract:There is a lack of benchmarks for evaluating large language models (LLMs) in long-form medical question answering (QA). Most existing medical QA evaluation benchmarks focus on automatic metrics and multiple-choice questions. While valuable, these benchmarks fail to fully capture or assess the complexities of real-world clinical applications where LLMs are being deployed. Furthermore, existing studies on evaluating long-form answer generation in medical QA are primarily closed-source, lacking access to human medical expert annotations, which makes it difficult to reproduce results and enhance existing baselines. In this work, we introduce a new publicly available benchmark featuring real-world consumer medical questions with long-form answer evaluations annotated by medical doctors. We performed pairwise comparisons of responses from various open and closed-source medical and general-purpose LLMs based on criteria such as correctness, helpfulness, harmfulness, and bias. Additionally, we performed a comprehensive LLM-as-a-judge analysis to study the alignment between human judgments and LLMs. Our preliminary results highlight the strong potential of open LLMs in medical QA compared to leading closed models. Code & Data: https://github.com/lavita-ai/medical-eval-sphere
A Petri Dish for Histopathology Image Analysis
Jan 29, 2021



Abstract:With the rise of deep learning, there has been increased interest in using neural networks for histopathology image analysis, a field that investigates the properties of biopsy or resected specimens that are traditionally manually examined under a microscope by pathologists. In histopathology image analysis, however, challenges such as limited data, costly annotation, and processing high-resolution and variable-size images create a high barrier of entry and make it difficult to quickly iterate over model designs. Throughout scientific history, many significant research directions have leveraged small-scale experimental setups as petri dishes to efficiently evaluate exploratory ideas, which are then validated in large-scale applications. For instance, the Drosophila fruit fly in genetics and MNIST in computer vision are well-known petri dishes. In this paper, we introduce a minimalist histopathology image analysis dataset (MHIST), an analogous petri dish for histopathology image analysis. MHIST is a binary classification dataset of 3,152 fixed-size images of colorectal polyps, each with a gold-standard label determined by the majority vote of seven board-certified gastrointestinal pathologists and annotator agreement level. MHIST occupies less than 400 MB of disk space, and a ResNet-18 baseline can be trained to convergence on MHIST in just 6 minutes using 3.5 GB of memory on a NVIDIA RTX 3090. As example use cases, we use MHIST to study natural questions such as how dataset size, network depth, transfer learning, and high-disagreement examples affect model performance. By introducing MHIST, we hope to not only help facilitate the work of current histopathology imaging researchers, but also make histopathology image analysis more accessible to the general computer vision community. Our dataset is available at https://bmirds.github.io/MHIST.
Development and Evaluation of a Deep Neural Network for Histologic Classification of Renal Cell Carcinoma on Biopsy and Surgical Resection Slides
Oct 30, 2020



Abstract:Renal cell carcinoma (RCC) is the most common renal cancer in adults. The histopathologic classification of RCC is essential for diagnosis, prognosis, and management of patients. Reorganization and classification of complex histologic patterns of RCC on biopsy and surgical resection slides under a microscope remains a heavily specialized, error-prone, and time-consuming task for pathologists. In this study, we developed a deep neural network model that can accurately classify digitized surgical resection slides and biopsy slides into five related classes: clear cell RCC, papillary RCC, chromophobe RCC, renal oncocytoma, and normal. In addition to the whole-slide classification pipeline, we visualized the identified indicative regions and features on slides for classification by reprocessing patch-level classification results to ensure the explainability of our diagnostic model. We evaluated our model on independent test sets of 78 surgical resection whole slides and 79 biopsy slides from our tertiary medical institution, and 69 randomly selected surgical resection slides from The Cancer Genome Atlas (TCGA) database. The average area under the curve (AUC) of our classifier on the internal resection slides, internal biopsy slides, and external TCGA slides is 0.98, 0.98 and 0.99, respectively. Our results suggest that the high generalizability of our approach across different data sources and specimen types. More importantly, our model has the potential to assist pathologists by (1) automatically pre-screening slides to reduce false-negative cases, (2) highlighting regions of importance on digitized slides to accelerate diagnosis, and (3) providing objective and accurate diagnosis as the second opinion.
Learn like a Pathologist: Curriculum Learning by Annotator Agreement for Histopathology Image Classification
Sep 29, 2020



Abstract:Applying curriculum learning requires both a range of difficulty in data and a method for determining the difficulty of examples. In many tasks, however, satisfying these requirements can be a formidable challenge. In this paper, we contend that histopathology image classification is a compelling use case for curriculum learning. Based on the nature of histopathology images, a range of difficulty inherently exists among examples, and, since medical datasets are often labeled by multiple annotators, annotator agreement can be used as a natural proxy for the difficulty of a given example. Hence, we propose a simple curriculum learning method that trains on progressively-harder images as determined by annotator agreement. We evaluate our hypothesis on the challenging and clinically-important task of colorectal polyp classification. Whereas vanilla training achieves an AUC of 83.7% for this task, a model trained with our proposed curriculum learning approach achieves an AUC of 88.2%, an improvement of 4.5%. Our work aims to inspire researchers to think more creatively and rigorously when choosing contexts for applying curriculum learning.
Difficulty Translation in Histopathology Images
Apr 27, 2020



Abstract:The unique nature of histopathology images opens the door to domain-specific formulations of image translation models. We propose a difficulty translation model that modifies colorectal histopathology images to become more challenging to classify. Our model comprises a scorer, which provides an output confidence to measure the difficulty of images, and an image translator, which learns to translate images from easy-to-classify to hard-to-classify using a training set defined by the scorer. We present three findings. First, generated images were indeed harder to classify for both human pathologists and machine learning classifiers than their corresponding source images. Second, image classifiers trained with generated images as augmented data performed better on both easy and hard images from an independent test set. Finally, human annotator agreement and our model's measure of difficulty correlated strongly, implying that for future work requiring human annotator agreement, the confidence score of a machine learning classifier could be used instead as a proxy.
Parallel Distributed Logistic Regression for Vertical Federated Learning without Third-Party Coordinator
Nov 22, 2019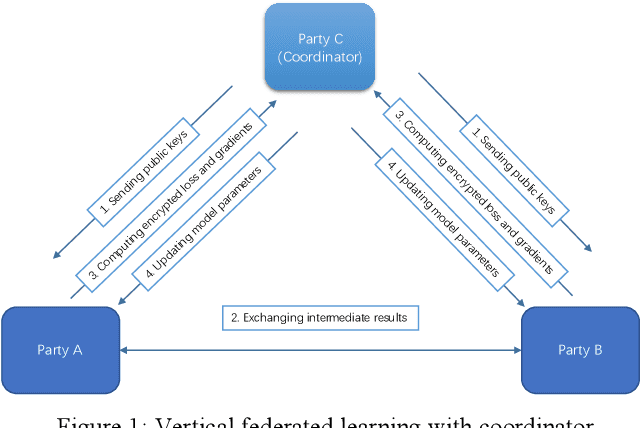
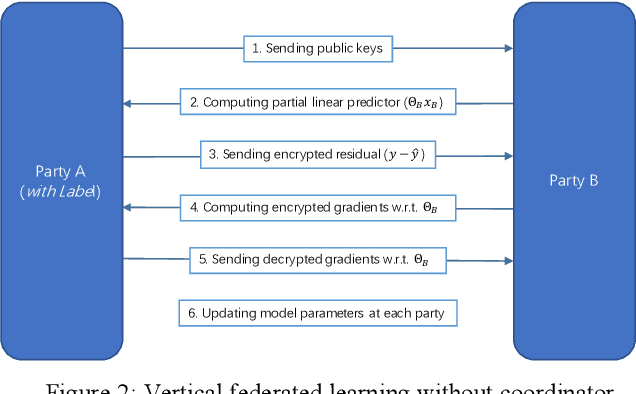
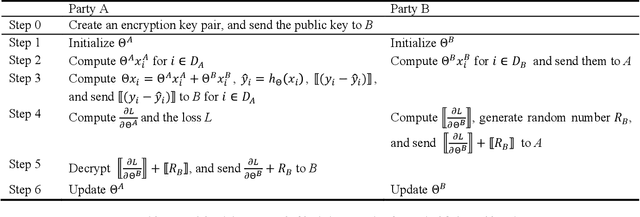
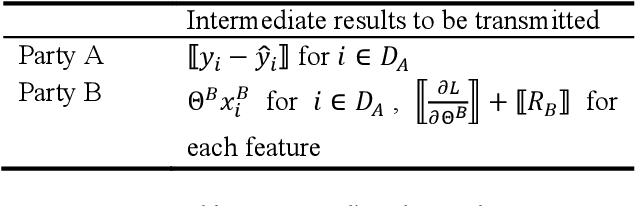
Abstract:Federated Learning is a new distributed learning mechanism which allows model training on a large corpus of decentralized data owned by different data providers, without sharing or leakage of raw data. According to the characteristics of data dis-tribution, it could be usually classified into three categories: horizontal federated learning, vertical federated learning, and federated transfer learning. In this paper we present a solution for parallel dis-tributed logistic regression for vertical federated learning. As compared with existing works, the role of third-party coordinator is removed in our proposed solution. The system is built on the pa-rameter server architecture and aims to speed up the model training via utilizing a cluster of servers in case of large volume of training data. We also evaluate the performance of the parallel distributed model training and the experimental results show the great scalability of the system.
Generative Image Translation for Data Augmentation in Colorectal Histopathology Images
Oct 13, 2019



Abstract:We present an image translation approach to generate augmented data for mitigating data imbalances in a dataset of histopathology images of colorectal polyps, adenomatous tumors that can lead to colorectal cancer if left untreated. By applying cycle-consistent generative adversarial networks (CycleGANs) to a source domain of normal colonic mucosa images, we generate synthetic colorectal polyp images that belong to diagnostically less common polyp classes. Generated images maintain the general structure of their source image but exhibit adenomatous features that can be enhanced with our proposed filtration module, called Path-Rank-Filter. We evaluate the quality of generated images through Turing tests with four gastrointestinal pathologists, finding that at least two of the four pathologists could not identify generated images at a statistically significant level. Finally, we demonstrate that using CycleGAN-generated images to augment training data improves the AUC of a convolutional neural network for detecting sessile serrated adenomas by over 10%, suggesting that our approach might warrant further research for other histopathology image classification tasks.
Deep neural networks for automated classification of colorectal polyps on histopathology slides: A multi-institutional evaluation
Sep 27, 2019



Abstract:Histological classification of colorectal polyps plays a critical role in both screening for colorectal cancer and care of affected patients. In this study, we developed a deep neural network for classification of four major colorectal polyp types on digitized histopathology slides and compared its performance to local pathologists' diagnoses at the point-of-care retrieved from corresponding pathology labs. We evaluated the deep neural network on an internal dataset of 157 histopathology slides from the Dartmouth-Hitchcock Medical Center (DHMC) in New Hampshire, as well as an external dataset of 513 histopathology slides from 24 different institutions spanning 13 states in the United States. For the internal evaluation, the deep neural network had a mean accuracy of 93.5% (95% CI 89.6%-97.4%), compared with local pathologists' accuracy of 91.4% (95% CI 87.0%-95.8%). On the external test set, the deep neural network achieved an accuracy of 85.7% (95% CI 82.7%-88.7%), significantly outperforming the accuracy of local pathologists at 80.9% (95% CI 77.5%-84.3%, p<0.05) at the point-of-care. If confirmed in clinical settings, our model could assist pathologists by improving the diagnostic efficiency, reproducibility, and accuracy of colorectal cancer screenings.
Automated detection of celiac disease on duodenal biopsy slides: a deep learning approach
Jan 31, 2019



Abstract:Celiac disease prevalence and diagnosis have increased substantially in recent years. The current gold standard for celiac disease confirmation is visual examination of duodenal mucosal biopsies. An accurate computer-aided biopsy analysis system using deep learning can help pathologists diagnose celiac disease more efficiently. In this study, we trained a deep learning model to detect celiac disease on duodenal biopsy images. Our model uses a state-of-the-art residual convolutional neural network to evaluate patches of duodenal tissue and then aggregates those predictions for whole-slide classification. We tested the model on an independent set of 212 images and evaluated its classification results against reference standards established by pathologists. Our model identified celiac disease, normal tissue, and nonspecific duodenitis with accuracies of 95.3%, 91.0%, and 89.2%, respectively. The area under the receiver operating characteristic curve was greater than 0.95 for all classes. We have developed an automated biopsy analysis system that achieves high performance in detecting celiac disease on biopsy slides. Our system can highlight areas of interest and provide preliminary classification of duodenal biopsies prior to review by pathologists. This technology has great potential for improving the accuracy and efficiency of celiac disease diagnosis.
Finding a Needle in the Haystack: Attention-Based Classification of High Resolution Microscopy Images
Nov 20, 2018
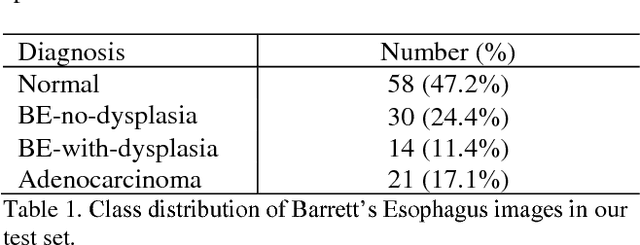
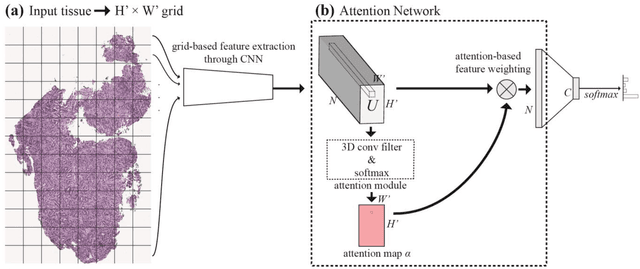
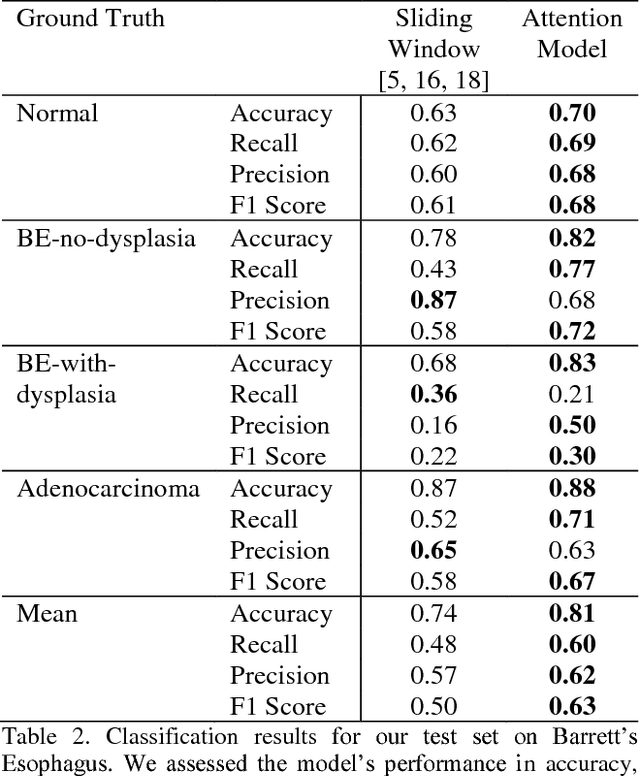
Abstract:Deep learning for classification of microscopy images is challenging because whole-slide images are high resolution. Due to the large size of these images, they cannot be transferred into GPU memory, so there are currently no end-to-end deep learning architectures for their analysis. Existing work has used a sliding window for crop classification, followed by a heuristic to determine the label for the whole slide. This pipeline is not efficient or robust, however, because crops are analyzed independently of their neighbors and the decisive features for classifying a whole slide are only found in a few regions of interest. In this paper, we present an attention-based model for classification of high resolution microscopy images. Our model dynamically finds regions of interest from a wide-view, then identifies characteristic patterns in those regions for whole-slide classification. This approach is analogous to how pathologists examine slides under the microscope and is the first to generalize the attention mechanism to high resolution images. Furthermore, our model does not require bounding box annotations for the regions of interest and is trainable end-to-end with flexible input. We evaluated our model on a microscopy dataset of Barrett's Esophagus images, and the results showed that our approach outperforms the current state-of-the-art sliding window method by a large margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge