Finding a Needle in the Haystack: Attention-Based Classification of High Resolution Microscopy Images
Paper and Code
Nov 20, 2018
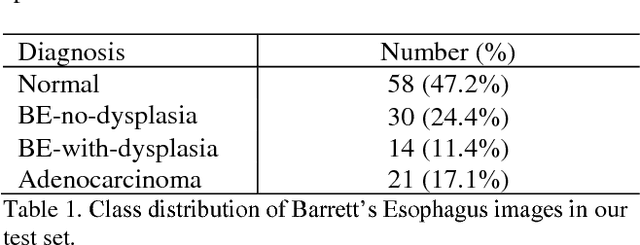
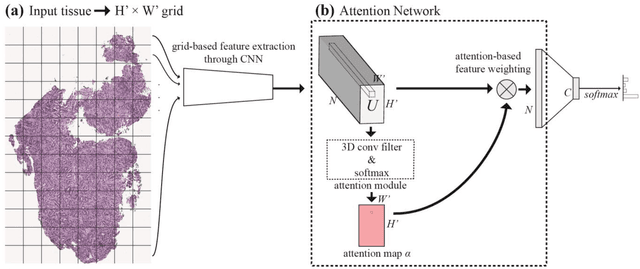
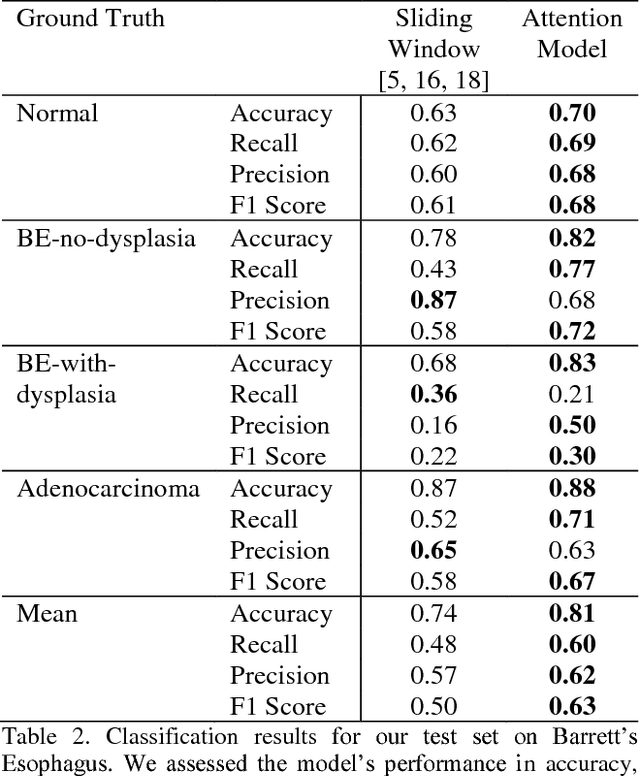
Deep learning for classification of microscopy images is challenging because whole-slide images are high resolution. Due to the large size of these images, they cannot be transferred into GPU memory, so there are currently no end-to-end deep learning architectures for their analysis. Existing work has used a sliding window for crop classification, followed by a heuristic to determine the label for the whole slide. This pipeline is not efficient or robust, however, because crops are analyzed independently of their neighbors and the decisive features for classifying a whole slide are only found in a few regions of interest. In this paper, we present an attention-based model for classification of high resolution microscopy images. Our model dynamically finds regions of interest from a wide-view, then identifies characteristic patterns in those regions for whole-slide classification. This approach is analogous to how pathologists examine slides under the microscope and is the first to generalize the attention mechanism to high resolution images. Furthermore, our model does not require bounding box annotations for the regions of interest and is trainable end-to-end with flexible input. We evaluated our model on a microscopy dataset of Barrett's Esophagus images, and the results showed that our approach outperforms the current state-of-the-art sliding window method by a large margin.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge