Arief A. Suriawinata
Masked Pre-Training of Transformers for Histology Image Analysis
Apr 14, 2023Abstract:In digital pathology, whole slide images (WSIs) are widely used for applications such as cancer diagnosis and prognosis prediction. Visual transformer models have recently emerged as a promising method for encoding large regions of WSIs while preserving spatial relationships among patches. However, due to the large number of model parameters and limited labeled data, applying transformer models to WSIs remains challenging. Inspired by masked language models, we propose a pretext task for training the transformer model without labeled data to address this problem. Our model, MaskHIT, uses the transformer output to reconstruct masked patches and learn representative histological features based on their positions and visual features. The experimental results demonstrate that MaskHIT surpasses various multiple instance learning approaches by 3% and 2% on survival prediction and cancer subtype classification tasks, respectively. Furthermore, MaskHIT also outperforms two of the most recent state-of-the-art transformer-based methods. Finally, a comparison between the attention maps generated by the MaskHIT model with pathologist's annotations indicates that the model can accurately identify clinically relevant histological structures in each task.
MHAttnSurv: Multi-Head Attention for Survival Prediction Using Whole-Slide Pathology Images
Oct 22, 2021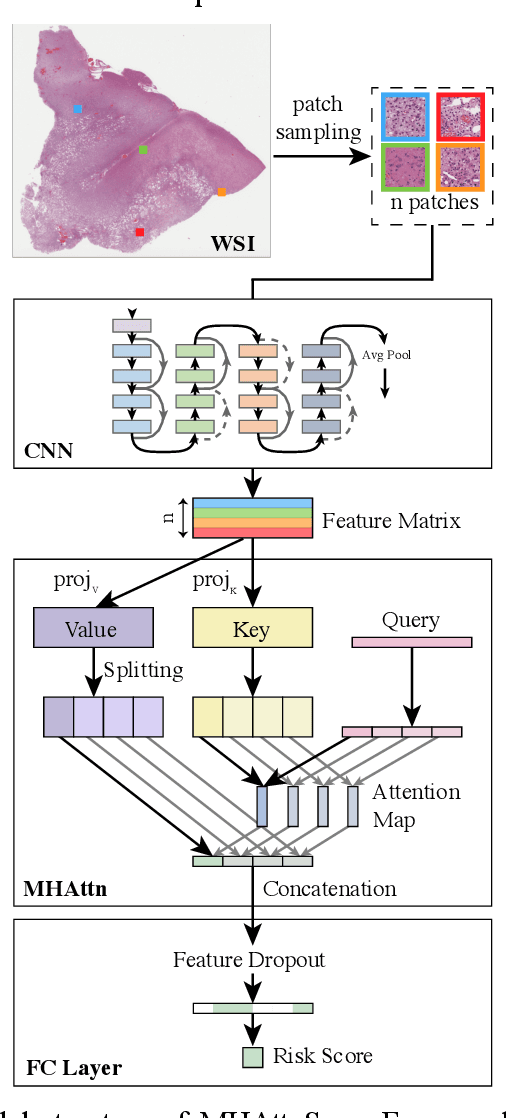
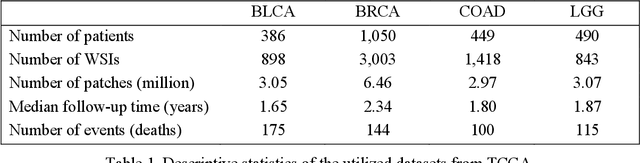

Abstract:In pathology, whole-slide images (WSI) based survival prediction has attracted increasing interest. However, given the large size of WSIs and the lack of pathologist annotations, extracting the prognostic information from WSIs remains a challenging task. Previous studies have used multiple instance learning approaches to combine the information from multiple randomly sampled patches, but different visual patterns may contribute differently to prognosis prediction. In this study, we developed a multi-head attention approach to focus on various parts of a tumor slide, for more comprehensive information extraction from WSIs. We evaluated our approach on four cancer types from The Cancer Genome Atlas database. Our model achieved an average c-index of 0.640, outperforming two existing state-of-the-art approaches for WSI-based survival prediction, which have an average c-index of 0.603 and 0.619 on these datasets. Visualization of our attention maps reveals each attention head focuses synergistically on different morphological patterns.
Resolution-Based Distillation for Efficient Histology Image Classification
Jan 11, 2021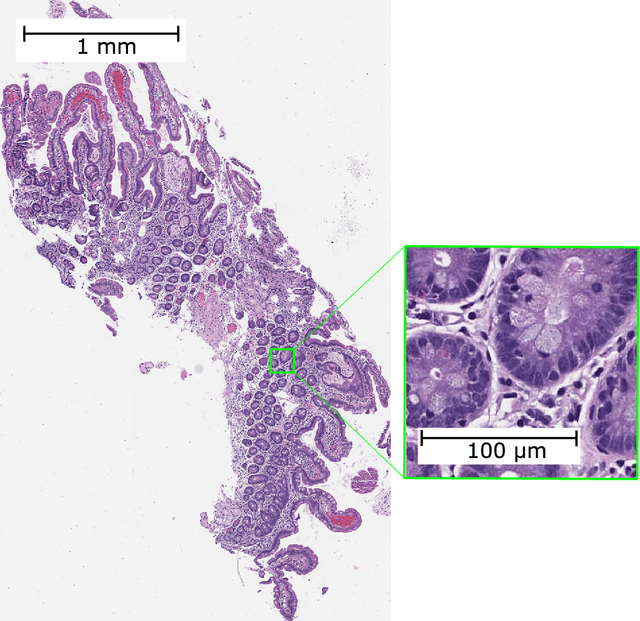
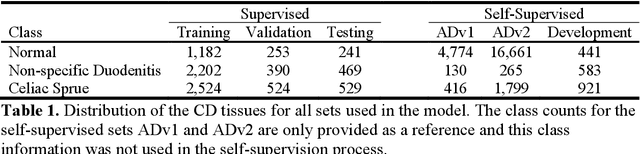
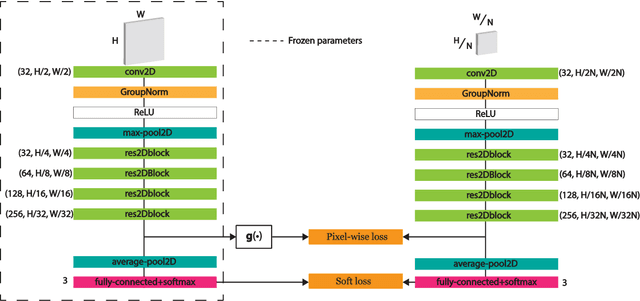
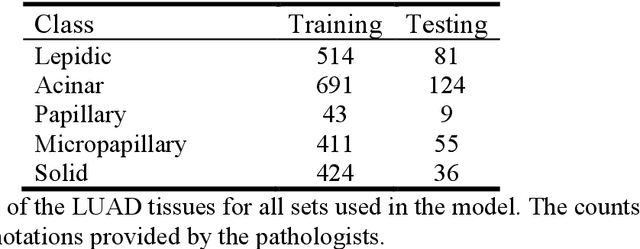
Abstract:Developing deep learning models to analyze histology images has been computationally challenging, as the massive size of the images causes excessive strain on all parts of the computing pipeline. This paper proposes a novel deep learning-based methodology for improving the computational efficiency of histology image classification. The proposed approach is robust when used with images that have reduced input resolution and can be trained effectively with limited labeled data. Pre-trained on the original high-resolution (HR) images, our method uses knowledge distillation (KD) to transfer learned knowledge from a teacher model to a student model trained on the same images at a much lower resolution. To address the lack of large-scale labeled histology image datasets, we perform KD in a self-supervised manner. We evaluate our approach on two histology image datasets associated with celiac disease (CD) and lung adenocarcinoma (LUAD). Our results show that a combination of KD and self-supervision allows the student model to approach, and in some cases, surpass the classification accuracy of the teacher, while being much more efficient. Additionally, we observe an increase in student classification performance as the size of the unlabeled dataset increases, indicating that there is potential to scale further. For the CD data, our model outperforms the HR teacher model, while needing 4 times fewer computations. For the LUAD data, our student model results at 1.25x magnification are within 3% of the teacher model at 10x magnification, with a 64 times computational cost reduction. Moreover, our CD outcomes benefit from performance scaling with the use of more unlabeled data. For 0.625x magnification, using unlabeled data improves accuracy by 4% over the baseline. Thus, our method can improve the feasibility of deep learning solutions for digital pathology with standard computational hardware.
Predicting colorectal polyp recurrence using time-to-event analysis of medical records
Nov 18, 2019



Abstract:Identifying patient characteristics that influence the rate of colorectal polyp recurrence can provide important insights into which patients are at higher risk for recurrence. We used natural language processing to extract polyp morphological characteristics from 953 polyp-presenting patients' electronic medical records. We used subsequent colonoscopy reports to examine how the time to polyp recurrence (731 patients experienced recurrence) is influenced by these characteristics as well as anthropometric features using Kaplan-Meier curves, Cox proportional hazards modeling, and random survival forest models. We found that the rate of recurrence differed significantly by polyp size, number, and location and patient smoking status. Additionally, right-sided colon polyps increased recurrence risk by 30% compared to left-sided polyps. History of tobacco use increased polyp recurrence risk by 20% compared to never-users. A random survival forest model showed an AUC of 0.65 and identified several other predictive variables, which can inform development of personalized polyp surveillance plans.
Deep neural networks for automated classification of colorectal polyps on histopathology slides: A multi-institutional evaluation
Sep 27, 2019



Abstract:Histological classification of colorectal polyps plays a critical role in both screening for colorectal cancer and care of affected patients. In this study, we developed a deep neural network for classification of four major colorectal polyp types on digitized histopathology slides and compared its performance to local pathologists' diagnoses at the point-of-care retrieved from corresponding pathology labs. We evaluated the deep neural network on an internal dataset of 157 histopathology slides from the Dartmouth-Hitchcock Medical Center (DHMC) in New Hampshire, as well as an external dataset of 513 histopathology slides from 24 different institutions spanning 13 states in the United States. For the internal evaluation, the deep neural network had a mean accuracy of 93.5% (95% CI 89.6%-97.4%), compared with local pathologists' accuracy of 91.4% (95% CI 87.0%-95.8%). On the external test set, the deep neural network achieved an accuracy of 85.7% (95% CI 82.7%-88.7%), significantly outperforming the accuracy of local pathologists at 80.9% (95% CI 77.5%-84.3%, p<0.05) at the point-of-care. If confirmed in clinical settings, our model could assist pathologists by improving the diagnostic efficiency, reproducibility, and accuracy of colorectal cancer screenings.
Automated detection of celiac disease on duodenal biopsy slides: a deep learning approach
Jan 31, 2019



Abstract:Celiac disease prevalence and diagnosis have increased substantially in recent years. The current gold standard for celiac disease confirmation is visual examination of duodenal mucosal biopsies. An accurate computer-aided biopsy analysis system using deep learning can help pathologists diagnose celiac disease more efficiently. In this study, we trained a deep learning model to detect celiac disease on duodenal biopsy images. Our model uses a state-of-the-art residual convolutional neural network to evaluate patches of duodenal tissue and then aggregates those predictions for whole-slide classification. We tested the model on an independent set of 212 images and evaluated its classification results against reference standards established by pathologists. Our model identified celiac disease, normal tissue, and nonspecific duodenitis with accuracies of 95.3%, 91.0%, and 89.2%, respectively. The area under the receiver operating characteristic curve was greater than 0.95 for all classes. We have developed an automated biopsy analysis system that achieves high performance in detecting celiac disease on biopsy slides. Our system can highlight areas of interest and provide preliminary classification of duodenal biopsies prior to review by pathologists. This technology has great potential for improving the accuracy and efficiency of celiac disease diagnosis.
Deep-Learning for Classification of Colorectal Polyps on Whole-Slide Images
Apr 12, 2017
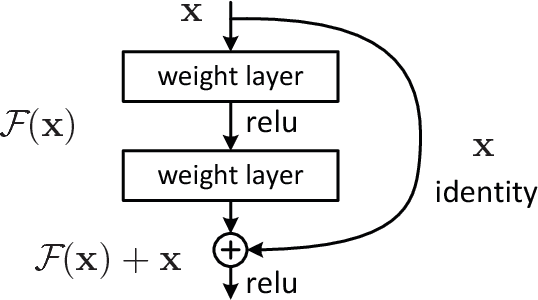
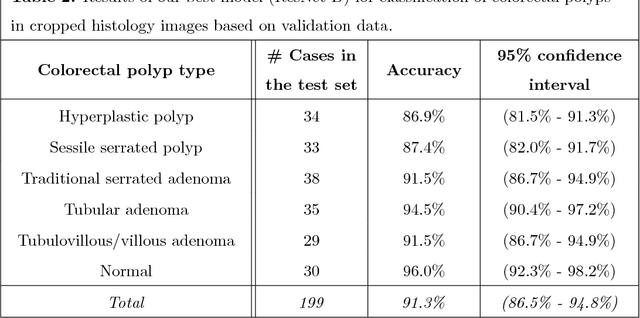
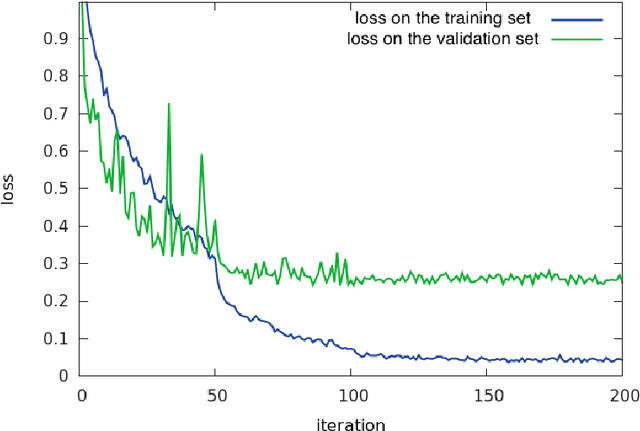
Abstract:Histopathological characterization of colorectal polyps is an important principle for determining the risk of colorectal cancer and future rates of surveillance for patients. This characterization is time-intensive, requires years of specialized training, and suffers from significant inter-observer and intra-observer variability. In this work, we built an automatic image-understanding method that can accurately classify different types of colorectal polyps in whole-slide histology images to help pathologists with histopathological characterization and diagnosis of colorectal polyps. The proposed image-understanding method is based on deep-learning techniques, which rely on numerous levels of abstraction for data representation and have shown state-of-the-art results for various image analysis tasks. Our image-understanding method covers all five polyp types (hyperplastic polyp, sessile serrated polyp, traditional serrated adenoma, tubular adenoma, and tubulovillous/villous adenoma) that are included in the US multi-society task force guidelines for colorectal cancer risk assessment and surveillance, and encompasses the most common occurrences of colorectal polyps. Our evaluation on 239 independent test samples shows our proposed method can identify the types of colorectal polyps in whole-slide images with a high efficacy (accuracy: 93.0%, precision: 89.7%, recall: 88.3%, F1 score: 88.8%). The presented method in this paper can reduce the cognitive burden on pathologists and improve their accuracy and efficiency in histopathological characterization of colorectal polyps, and in subsequent risk assessment and follow-up recommendations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge