Laura J. Tafe
Vision Transformer-Based Deep Learning for Histologic Classification of Endometrial Cancer
Dec 13, 2023Abstract:Endometrial cancer, the sixth most common cancer in females worldwide, presents as a heterogeneous group with certain types prone to recurrence. Precise histologic evaluation of endometrial cancer is essential for effective patient management and determining the best treatment modalities. This study introduces EndoNet, a transformer-based deep learning approach for histologic classification of endometrial cancer. EndoNet uses convolutional neural networks for extracting histologic features and a vision transformer for aggregating these features and classifying slides based on their visual characteristics. The model was trained on 929 digitized hematoxylin and eosin-stained whole slide images of endometrial cancer from hysterectomy cases at Dartmouth Health. It classifies these slides into low grade (Endometroid Grades 1 and 2) and high-grade (endometroid carcinoma FIGO grade 3, uterine serous carcinoma, carcinosarcoma) categories. EndoNet was evaluated on an internal test set of 218 slides and an external test set of 100 random slides from the public TCGA database. The model achieved a weighted average F1-score of 0.92 (95% CI: 0.87-0.95) and an AUC of 0.93 (95% CI: 0.88-0.96) on the internal test, and 0.86 (95% CI: 0.80-0.94) for F1-score and 0.86 (95% CI: 0.75-0.93) for AUC on the external test. Pending further validation, EndoNet has the potential to assist pathologists in classifying challenging gynecologic pathology tumors and enhancing patient care.
Fluorescence molecular optomic signatures improve identification of tumors in head and neck specimens
Aug 29, 2022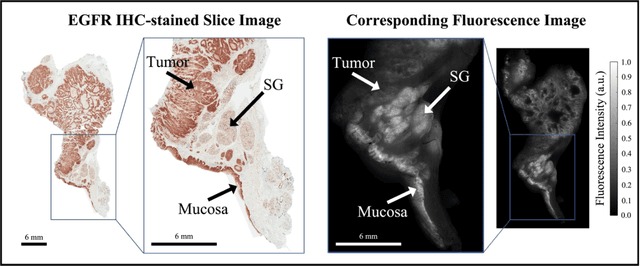
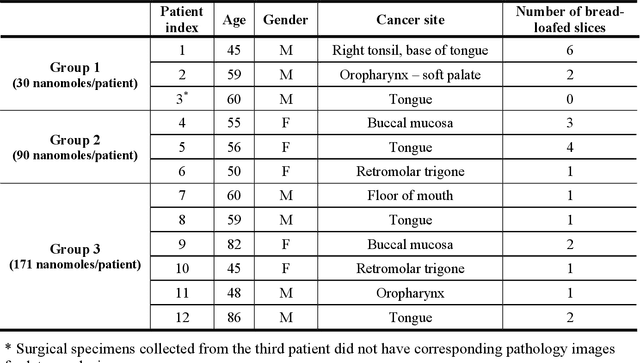
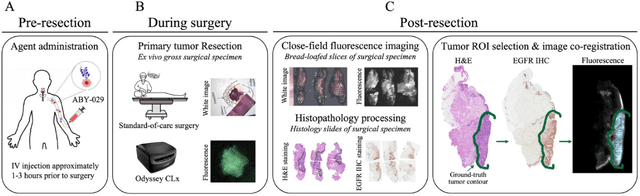
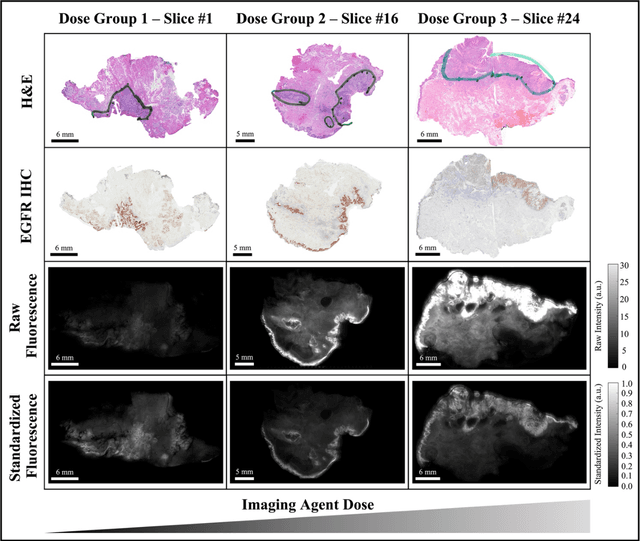
Abstract:In this study, a radiomics approach was extended to optical fluorescence molecular imaging data for tissue classification, termed 'optomics'. Fluorescence molecular imaging is emerging for precise surgical guidance during head and neck squamous cell carcinoma (HNSCC) resection. However, the tumor-to-normal tissue contrast is confounded by intrinsic physiological limitations of heterogeneous expression of the target molecule, epidermal growth factor receptor (EGFR). Optomics seek to improve tumor identification by probing textural pattern differences in EGFR expression conveyed by fluorescence. A total of 1,472 standardized optomic features were extracted from fluorescence image samples. A supervised machine learning pipeline involving a support vector machine classifier was trained with 25 top-ranked features selected by minimum redundancy maximum relevance criterion. Model predictive performance was compared to fluorescence intensity thresholding method by classifying testing set image patches of resected tissue with histologically confirmed malignancy status. The optomics approach provided consistent improvement in prediction accuracy on all test set samples, irrespective of dose, compared to fluorescence intensity thresholding method (mean accuracies of 89% vs. 81%; P = 0.0072). The improved performance demonstrates that extending the radiomics approach to fluorescence molecular imaging data offers a promising image analysis technique for cancer detection in fluorescence-guided surgery.
Resolution-Based Distillation for Efficient Histology Image Classification
Jan 11, 2021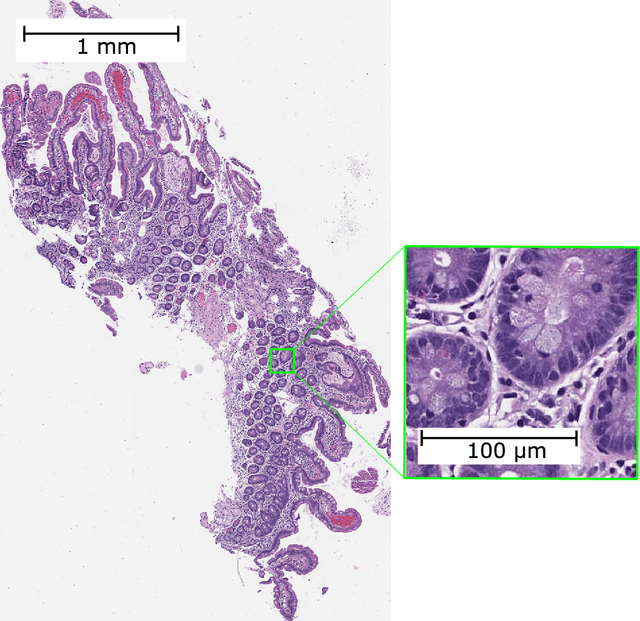
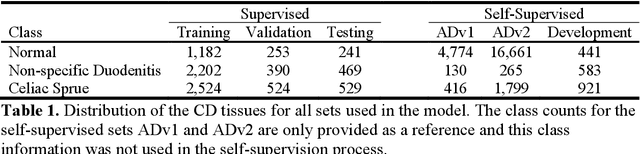
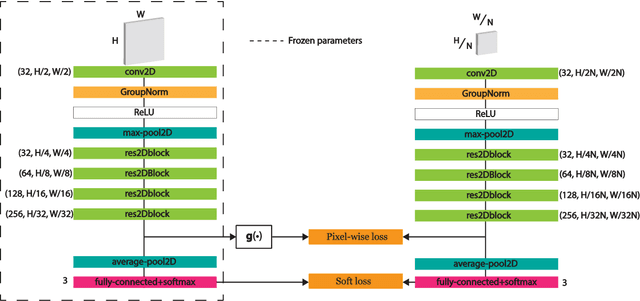
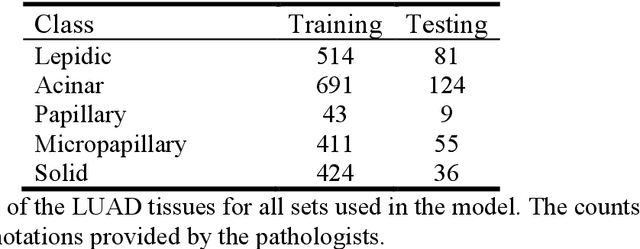
Abstract:Developing deep learning models to analyze histology images has been computationally challenging, as the massive size of the images causes excessive strain on all parts of the computing pipeline. This paper proposes a novel deep learning-based methodology for improving the computational efficiency of histology image classification. The proposed approach is robust when used with images that have reduced input resolution and can be trained effectively with limited labeled data. Pre-trained on the original high-resolution (HR) images, our method uses knowledge distillation (KD) to transfer learned knowledge from a teacher model to a student model trained on the same images at a much lower resolution. To address the lack of large-scale labeled histology image datasets, we perform KD in a self-supervised manner. We evaluate our approach on two histology image datasets associated with celiac disease (CD) and lung adenocarcinoma (LUAD). Our results show that a combination of KD and self-supervision allows the student model to approach, and in some cases, surpass the classification accuracy of the teacher, while being much more efficient. Additionally, we observe an increase in student classification performance as the size of the unlabeled dataset increases, indicating that there is potential to scale further. For the CD data, our model outperforms the HR teacher model, while needing 4 times fewer computations. For the LUAD data, our student model results at 1.25x magnification are within 3% of the teacher model at 10x magnification, with a 64 times computational cost reduction. Moreover, our CD outcomes benefit from performance scaling with the use of more unlabeled data. For 0.625x magnification, using unlabeled data improves accuracy by 4% over the baseline. Thus, our method can improve the feasibility of deep learning solutions for digital pathology with standard computational hardware.
Pathologist-level classification of histologic patterns on resected lung adenocarcinoma slides with deep neural networks
Jan 31, 2019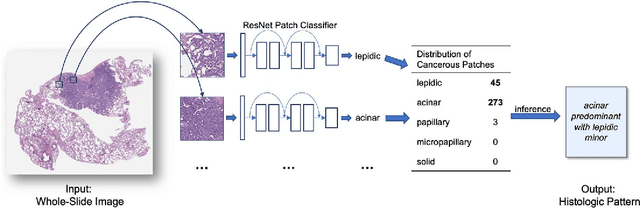
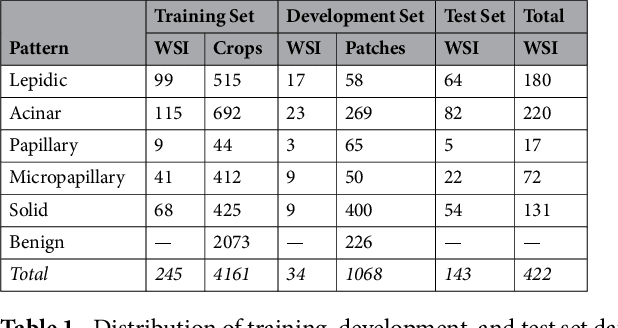
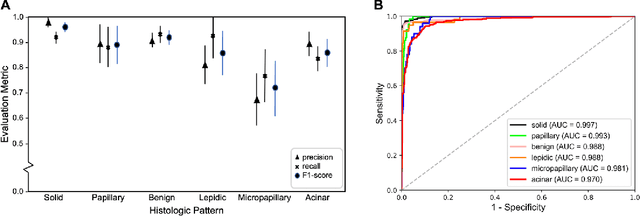
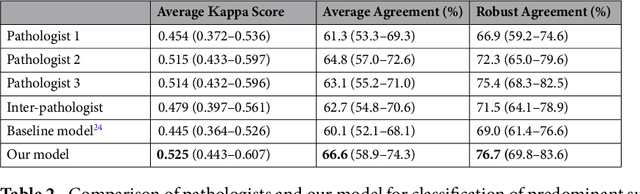
Abstract:Classification of histologic patterns in lung adenocarcinoma is critical for determining tumor grade and treatment for patients. However, this task is often challenging due to the heterogeneous nature of lung adenocarcinoma and the subjective criteria for evaluation. In this study, we propose a deep learning model that automatically classifies the histologic patterns of lung adenocarcinoma on surgical resection slides. Our model uses a convolutional neural network to identify regions of neoplastic cells, then aggregates those classifications to infer predominant and minor histologic patterns for any given whole-slide image. We evaluated our model on an independent set of 143 whole-slide images. It achieved a kappa score of 0.525 and an agreement of 66.6% with three pathologists for classifying the predominant patterns, slightly higher than the inter-pathologist kappa score of 0.485 and agreement of 62.7% on this test set. All evaluation metrics for our model and the three pathologists were within 95% confidence intervals of agreement. If confirmed in clinical practice, our model can assist pathologists in improving classification of lung adenocarcinoma patterns by automatically pre-screening and highlighting cancerous regions prior to review. Our approach can be generalized to any whole-slide image classification task, and code is made publicly available at https://github.com/BMIRDS/deepslide.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge