Kimberley S. Samkoe
Robust Real-time Segmentation of Bio-Morphological Features in Human Cherenkov Imaging during Radiotherapy via Deep Learning
Sep 09, 2024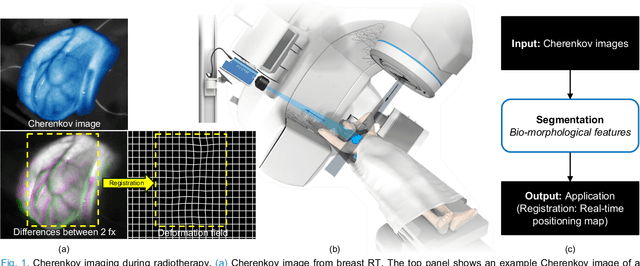
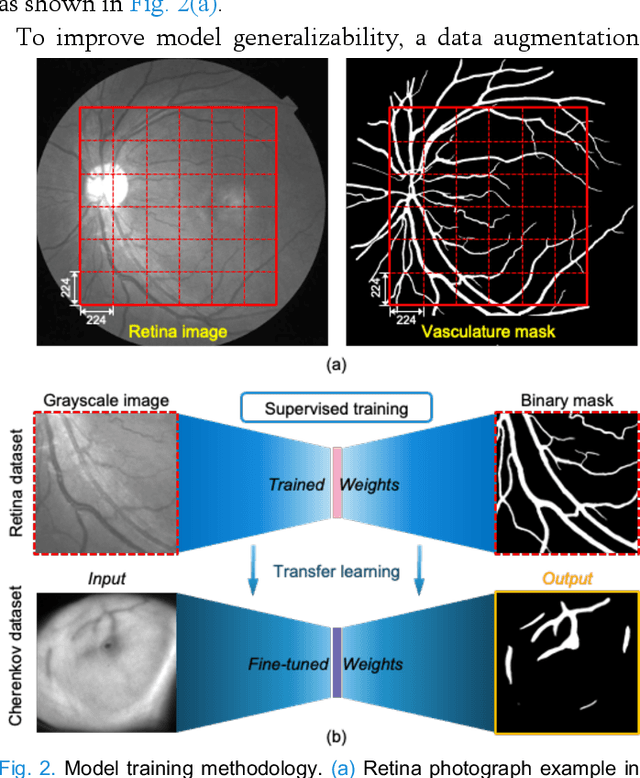

Abstract:Cherenkov imaging enables real-time visualization of megavoltage X-ray or electron beam delivery to the patient during Radiation Therapy (RT). Bio-morphological features, such as vasculature, seen in these images are patient-specific signatures that can be used for verification of positioning and motion management that are essential to precise RT treatment. However until now, no concerted analysis of this biological feature-based tracking was utilized because of the slow speed and accuracy of conventional image processing for feature segmentation. This study demonstrated the first deep learning framework for such an application, achieving video frame rate processing. To address the challenge of limited annotation of these features in Cherenkov images, a transfer learning strategy was applied. A fundus photography dataset including 20,529 patch retina images with ground-truth vessel annotation was used to pre-train a ResNet segmentation framework. Subsequently, a small Cherenkov dataset (1,483 images from 212 treatment fractions of 19 breast cancer patients) with known annotated vasculature masks was used to fine-tune the model for accurate segmentation prediction. This deep learning framework achieved consistent and rapid segmentation of Cherenkov-imaged bio-morphological features on another 19 patients, including subcutaneous veins, scars, and pigmented skin. Average segmentation by the model achieved Dice score of 0.85 and required less than 0.7 milliseconds processing time per instance. The model demonstrated outstanding consistency against input image variances and speed compared to conventional manual segmentation methods, laying the foundation for online segmentation in real-time monitoring in a prospective setting.
Cherenkov Imaged Bio-morphological Features Verify Patient Positioning with Deformable Tissue Translocation in Breast Radiotherapy
Sep 09, 2024Abstract:Accurate patient positioning is critical for precise radiotherapy dose delivery, as positioning errors can significantly affect treatment outcomes. This study introduces a novel method for tracking loco-regional tissue deformation through Cherenkov image analysis during fractionated breast cancer radiotherapy. The primary goal was to develop and test an algorithm for Cherenkov-based regional position accuracy quantification, specifically for loco-regional deformations, which lack ideal quantification methods in radiotherapy. Blood vessel detection and segmentation were developed in Cherenkov images using a tissue phantom with incremental movements, and later applied to images from fractionated whole breast radiotherapy in human patients (n=10). A combined rigid and non-rigid registration technique was used to detect inter- and intra-fractional positioning variations. This approach quantified positioning variations in two parts: a global shift from rigid registration and a two-dimensional variation map of loco-regional deformation from non-rigid registration. The methodology was validated using an anthropomorphic chest phantom experiment, where known treatment couch translations and respiratory motion were simulated to assess inter- and intra-fractional uncertainties, yielding an average accuracy of 0.83 mm for couch translations up to 20 mm. Analysis of clinical Cherenkov data from ten breast cancer patients showed an inter-fraction setup variation of 3.7 plus minus 2.4 mm relative to the first fraction and loco-regional deformations (95th percentile) of up to 3.3 plus minus 1.9 mm. This study presents a Cherenkov-based approach to quantify global and local positioning variations, demonstrating feasibility in addressing loco-regional deformations that conventional imaging techniques fail to capture.
Fluorescence molecular optomic signatures improve identification of tumors in head and neck specimens
Aug 29, 2022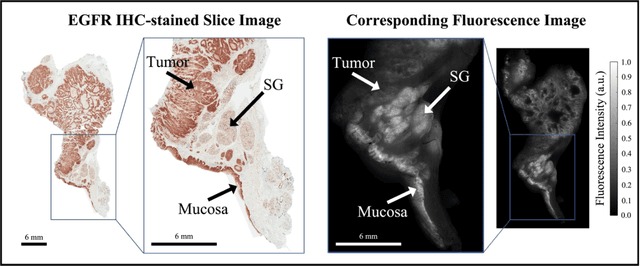
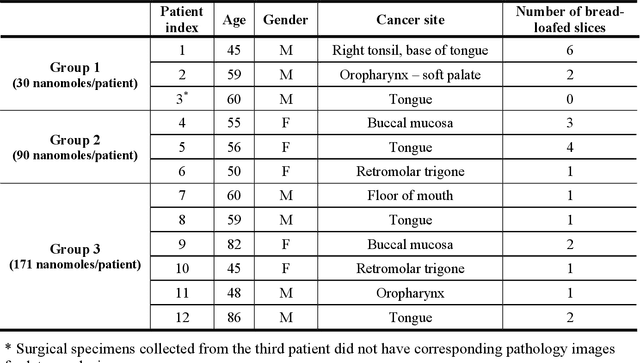
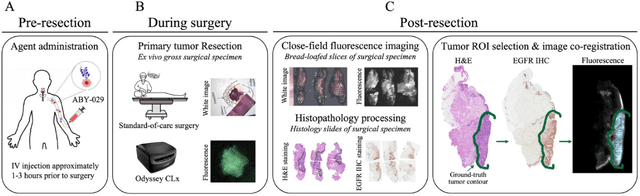
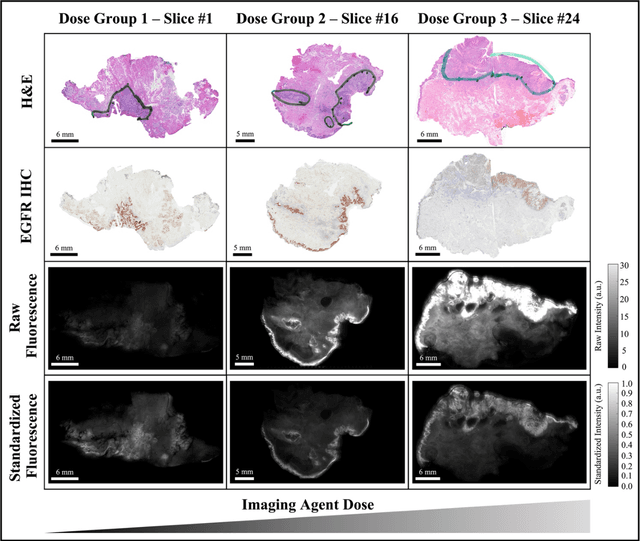
Abstract:In this study, a radiomics approach was extended to optical fluorescence molecular imaging data for tissue classification, termed 'optomics'. Fluorescence molecular imaging is emerging for precise surgical guidance during head and neck squamous cell carcinoma (HNSCC) resection. However, the tumor-to-normal tissue contrast is confounded by intrinsic physiological limitations of heterogeneous expression of the target molecule, epidermal growth factor receptor (EGFR). Optomics seek to improve tumor identification by probing textural pattern differences in EGFR expression conveyed by fluorescence. A total of 1,472 standardized optomic features were extracted from fluorescence image samples. A supervised machine learning pipeline involving a support vector machine classifier was trained with 25 top-ranked features selected by minimum redundancy maximum relevance criterion. Model predictive performance was compared to fluorescence intensity thresholding method by classifying testing set image patches of resected tissue with histologically confirmed malignancy status. The optomics approach provided consistent improvement in prediction accuracy on all test set samples, irrespective of dose, compared to fluorescence intensity thresholding method (mean accuracies of 89% vs. 81%; P = 0.0072). The improved performance demonstrates that extending the radiomics approach to fluorescence molecular imaging data offers a promising image analysis technique for cancer detection in fluorescence-guided surgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge