Manu Goyal
Prediction of Breast Cancer Recurrence Risk Using a Multi-Model Approach Integrating Whole Slide Imaging and Clinicopathologic Features
Jan 28, 2024Abstract:Breast cancer is the most common malignancy affecting women worldwide and is notable for its morphologic and biologic diversity, with varying risks of recurrence following treatment. The Oncotype DX Breast Recurrence Score test is an important predictive and prognostic genomic assay for estrogen receptor-positive breast cancer that guides therapeutic strategies; however, such tests can be expensive, delay care, and are not widely available. The aim of this study was to develop a multi-model approach integrating the analysis of whole slide images and clinicopathologic data to predict their associated breast cancer recurrence risks and categorize these patients into two risk groups according to the predicted score: low and high risk. The proposed novel methodology uses convolutional neural networks for feature extraction and vision transformers for contextual aggregation, complemented by a logistic regression model that analyzes clinicopathologic data for classification into two risk categories. This method was trained and tested on 993 hematoxylin and eosin-stained whole-slide images of breast cancers with corresponding clinicopathological features that had prior Oncotype DX testing. The model's performance was evaluated using an internal test set of 198 patients from Dartmouth Health and an external test set of 418 patients from the University of Chicago. The multi-model approach achieved an AUC of 0.92 (95 percent CI: 0.88-0.96) on the internal set and an AUC of 0.85 (95 percent CI: 0.79-0.90) on the external cohort. These results suggest that with further validation, the proposed methodology could provide an alternative to assist clinicians in personalizing treatment for breast cancer patients and potentially improving their outcomes.
Vision Transformer-Based Deep Learning for Histologic Classification of Endometrial Cancer
Dec 13, 2023Abstract:Endometrial cancer, the sixth most common cancer in females worldwide, presents as a heterogeneous group with certain types prone to recurrence. Precise histologic evaluation of endometrial cancer is essential for effective patient management and determining the best treatment modalities. This study introduces EndoNet, a transformer-based deep learning approach for histologic classification of endometrial cancer. EndoNet uses convolutional neural networks for extracting histologic features and a vision transformer for aggregating these features and classifying slides based on their visual characteristics. The model was trained on 929 digitized hematoxylin and eosin-stained whole slide images of endometrial cancer from hysterectomy cases at Dartmouth Health. It classifies these slides into low grade (Endometroid Grades 1 and 2) and high-grade (endometroid carcinoma FIGO grade 3, uterine serous carcinoma, carcinosarcoma) categories. EndoNet was evaluated on an internal test set of 218 slides and an external test set of 100 random slides from the public TCGA database. The model achieved a weighted average F1-score of 0.92 (95% CI: 0.87-0.95) and an AUC of 0.93 (95% CI: 0.88-0.96) on the internal test, and 0.86 (95% CI: 0.80-0.94) for F1-score and 0.86 (95% CI: 0.75-0.93) for AUC on the external test. Pending further validation, EndoNet has the potential to assist pathologists in classifying challenging gynecologic pathology tumors and enhancing patient care.
DiffGEPCI: 3D MRI Synthesis from mGRE Signals using 2.5D Diffusion Model
Nov 29, 2023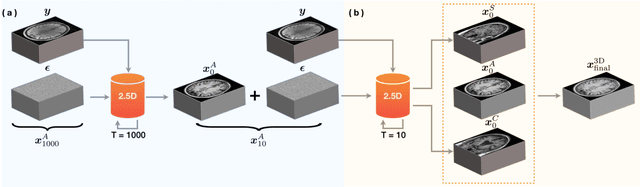
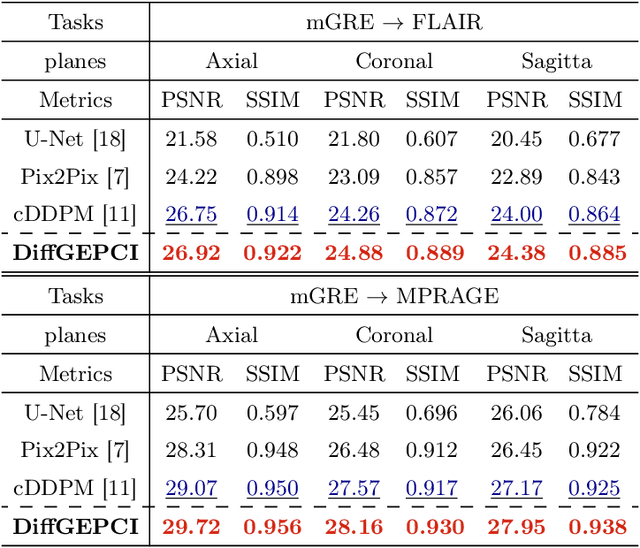
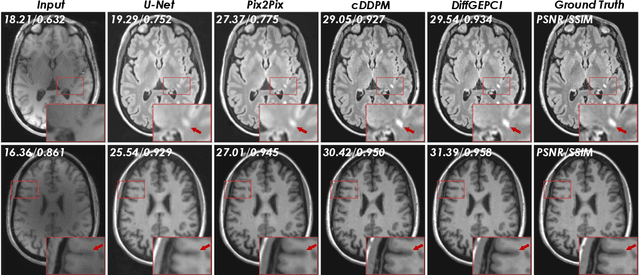
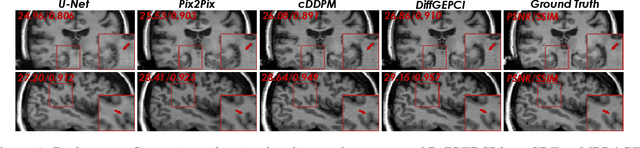
Abstract:We introduce a new framework called DiffGEPCI for cross-modality generation in magnetic resonance imaging (MRI) using a 2.5D conditional diffusion model. DiffGEPCI can synthesize high-quality Fluid Attenuated Inversion Recovery (FLAIR) and Magnetization Prepared-Rapid Gradient Echo (MPRAGE) images, without acquiring corresponding measurements, by leveraging multi-Gradient-Recalled Echo (mGRE) MRI signals as conditional inputs. DiffGEPCI operates in a two-step fashion: it initially estimates a 3D volume slice-by-slice using the axial plane and subsequently applies a refinement algorithm (referred to as 2.5D) to enhance the quality of the coronal and sagittal planes. Experimental validation on real mGRE data shows that DiffGEPCI achieves excellent performance, surpassing generative adversarial networks (GANs) and traditional diffusion models.
Development of Diabetic Foot Ulcer Datasets: An Overview
Jan 01, 2022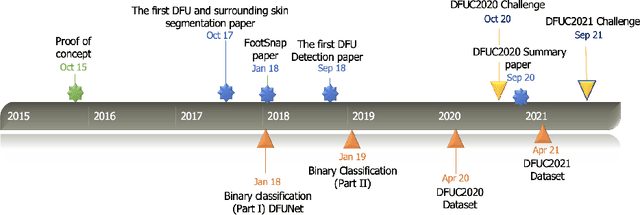

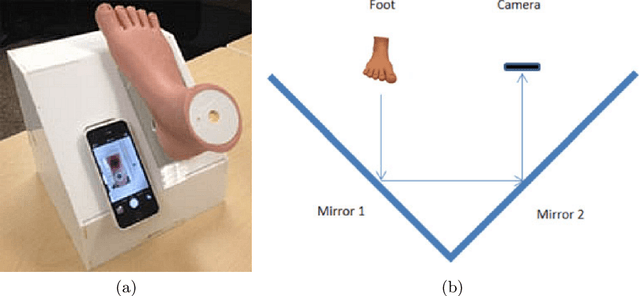
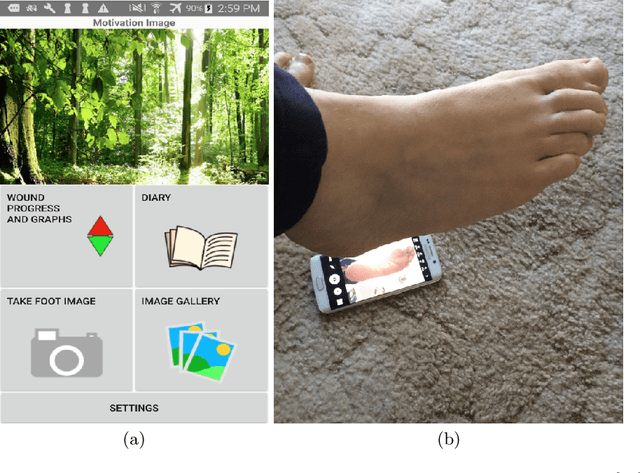
Abstract:This paper provides conceptual foundation and procedures used in the development of diabetic foot ulcer datasets over the past decade, with a timeline to demonstrate progress. We conduct a survey on data capturing methods for foot photographs, an overview of research in developing private and public datasets, the related computer vision tasks (detection, segmentation and classification), the diabetic foot ulcer challenges and the future direction of the development of the datasets. We report the distribution of dataset users by country and year. Our aim is to share the technical challenges that we encountered together with good practices in dataset development, and provide motivation for other researchers to participate in data sharing in this domain.
Automated Kidney Segmentation by Mask R-CNN in T2-weighted Magnetic Resonance Imaging
Aug 27, 2021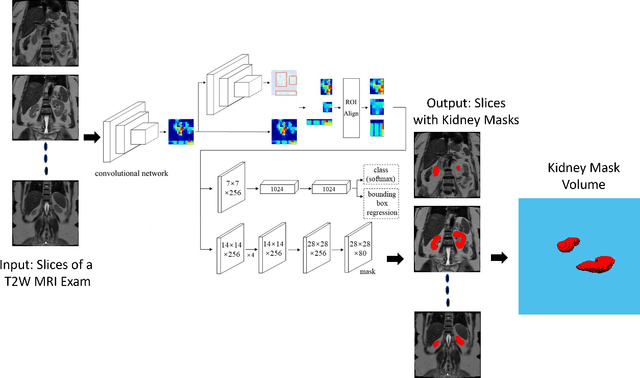
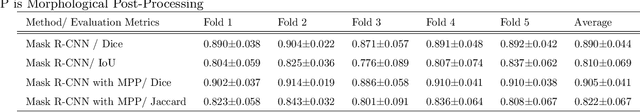
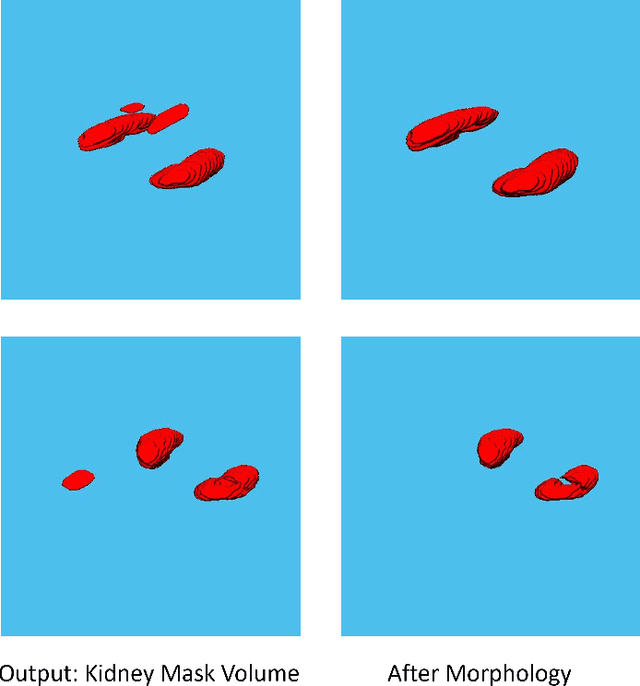
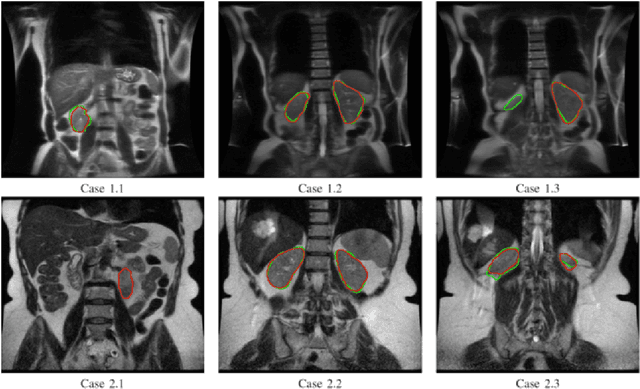
Abstract:Despite the recent advances of deep learning algorithms in medical imaging, the automatic segmentation algorithms for kidneys in MRI exams are still scarce. Automated segmentation of kidneys in Magnetic Resonance Imaging (MRI) exams are important for enabling radiomics and machine learning analysis of renal disease. In this work, we propose to use the popular Mask R-CNN for the automatic segmentation of kidneys in coronal T2-weighted Fast Spin Eco slices of 100 MRI exams. We propose the morphological operations as post-processing to further improve the performance of Mask R-CNN for this task. With 5-fold cross-validation data, the proposed Mask R-CNN is trained and validated on 70 and 10 MRI exams and then evaluated on the remaining 20 exams in each fold. Our proposed method achieved a dice score of 0.904 and IoU of 0.822.
Sensitivity and Specificity Evaluation of Deep Learning Models for Detection of Pneumoperitoneum on Chest Radiographs
Oct 17, 2020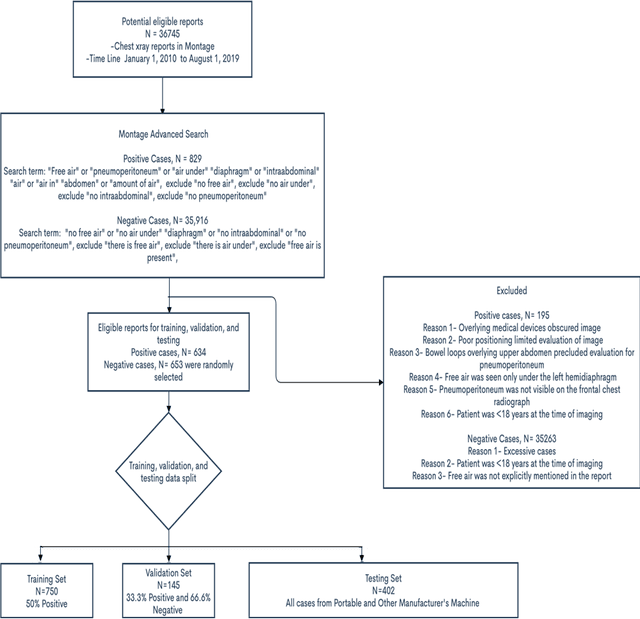
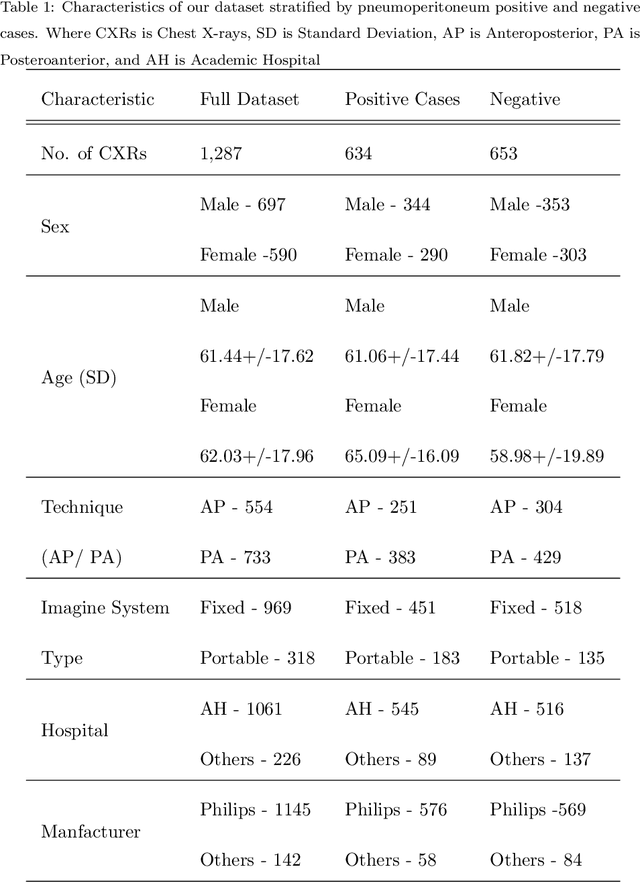
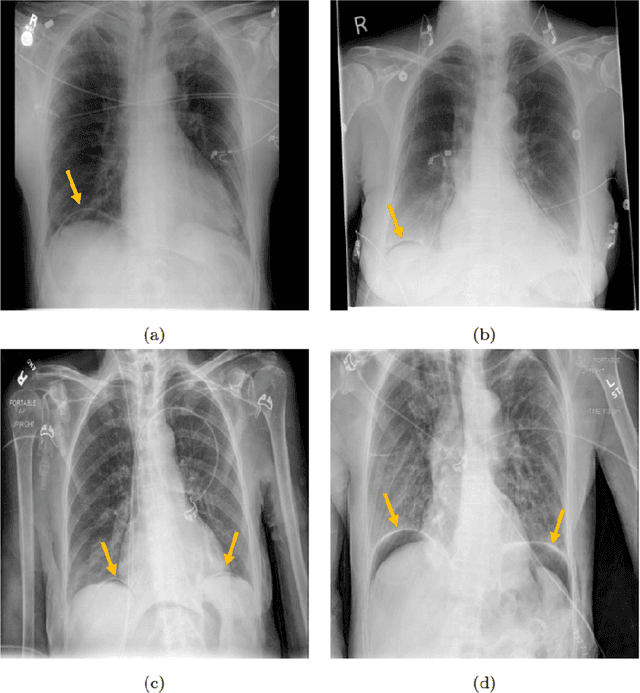
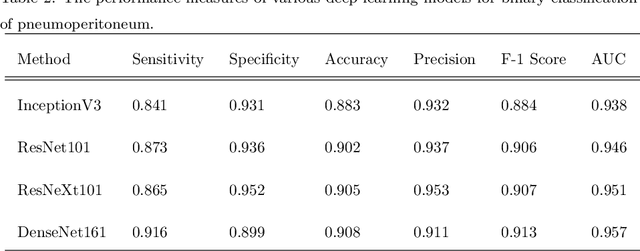
Abstract:Background: Deep learning has great potential to assist with detecting and triaging critical findings such as pneumoperitoneum on medical images. To be clinically useful, the performance of this technology still needs to be validated for generalizability across different types of imaging systems. Materials and Methods: This retrospective study included 1,287 chest X-ray images of patients who underwent initial chest radiography at 13 different hospitals between 2011 and 2019. The chest X-ray images were labelled independently by four radiologist experts as positive or negative for pneumoperitoneum. State-of-the-art deep learning models (ResNet101, InceptionV3, DenseNet161, and ResNeXt101) were trained on a subset of this dataset, and the automated classification performance was evaluated on the rest of the dataset by measuring the AUC, sensitivity, and specificity for each model. Furthermore, the generalizability of these deep learning models was assessed by stratifying the test dataset according to the type of the utilized imaging systems. Results: All deep learning models performed well for identifying radiographs with pneumoperitoneum, while DenseNet161 achieved the highest AUC of 95.7%, Specificity of 89.9%, and Sensitivity of 91.6%. DenseNet161 model was able to accurately classify radiographs from different imaging systems (Accuracy: 90.8%), while it was trained on images captured from a specific imaging system from a single institution. This result suggests the generalizability of our model for learning salient features in chest X-ray images to detect pneumoperitoneum, independent of the imaging system.
Deep Learning in Diabetic Foot Ulcers Detection: A Comprehensive Evaluation
Oct 15, 2020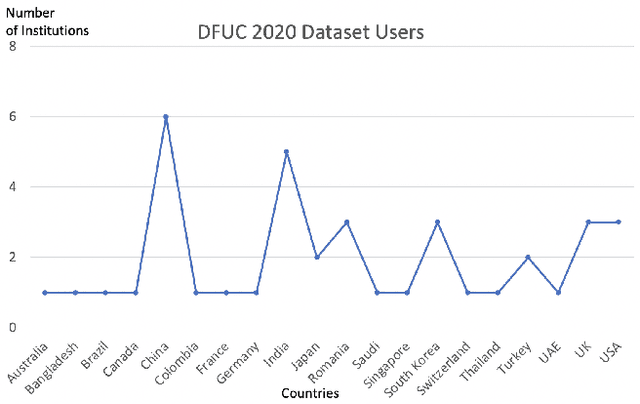
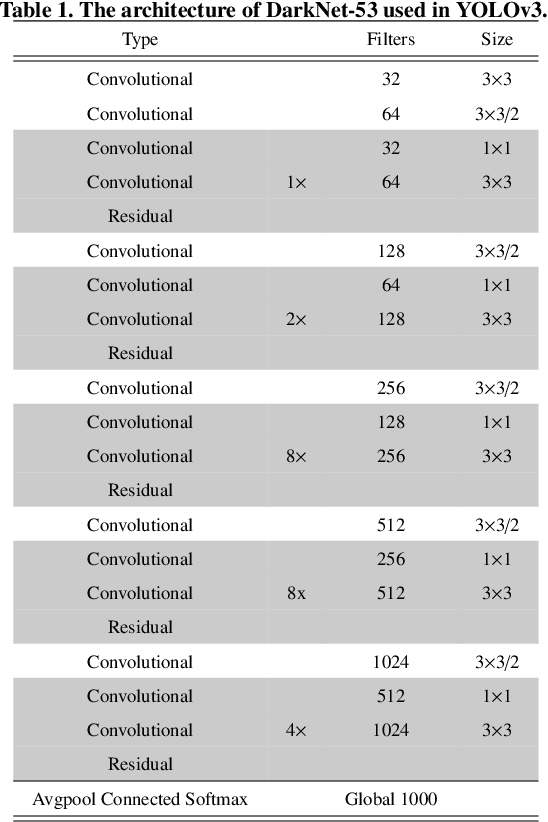
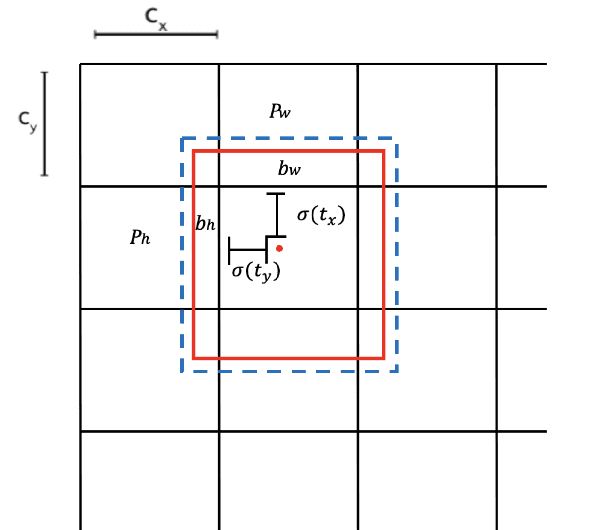
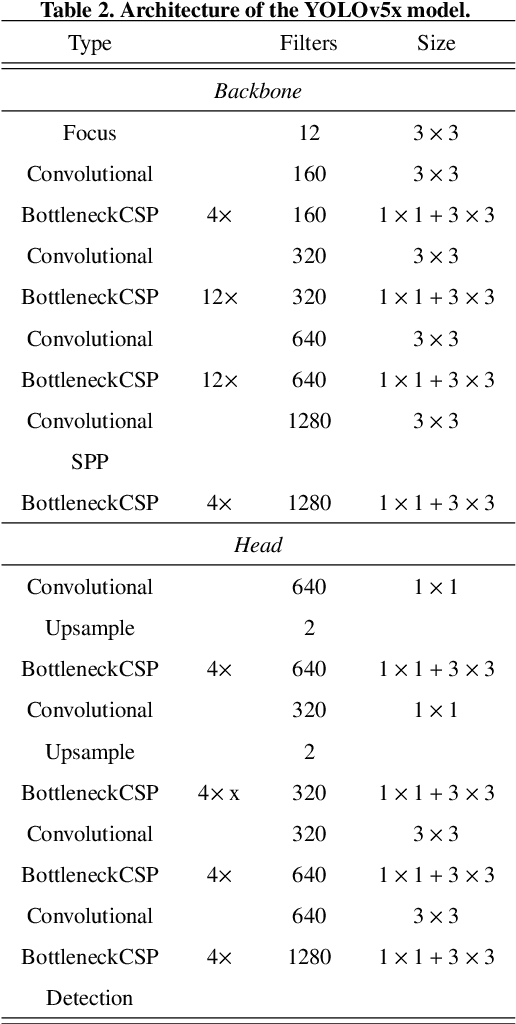
Abstract:There has been a substantial amount of research on computer methods and technology for the detection and recognition of diabetic foot ulcers (DFUs), but there is a lack of systematic comparisons of state-of-the-art deep learning object detection frameworks applied to this problem. With recent development and data sharing performed as part of the DFU Challenge (DFUC2020) such a comparison becomes possible: DFUC2020 provided participants with a comprehensive dataset consisting of 2,000 images for training each method and 2,000 images for testing them. The following deep learning-based algorithms are compared in this paper: Faster R-CNN, three variants of Faster R-CNN and an ensemble method; YOLOv3; YOLOv5; EfficientDet; and a new Cascade Attention Network. For each deep learning method, we provide a detailed description of model architecture, parameter settings for training and additional stages including pre-processing, data augmentation and post-processing. We provide a comprehensive evaluation for each method. All the methods required a data augmentation stage to increase the number of images available for training and a post-processing stage to remove false positives. The best performance is obtained Deformable Convolution, a variant of Faster R-CNN, with a mAP of 0.6940 and an F1-Score of 0.7434. Finally, we demonstrate that the ensemble method based on different deep learning methods can enhanced the F1-Score but not the mAP. Our results show that state-of-the-art deep learning methods can detect DFU with some accuracy, but there are many challenges ahead before they can be implemented in real world settings.
A Refined Deep Learning Architecture for Diabetic Foot Ulcers Detection
Jul 15, 2020



Abstract:Diabetic Foot Ulcers (DFU) that affect the lower extremities are a major complication of diabetes. Each year, more than 1 million diabetic patients undergo amputation due to failure to recognize DFU and get the proper treatment from clinicians. There is an urgent need to use a CAD system for the detection of DFU. In this paper, we propose using deep learning methods (EfficientDet Architectures) for the detection of DFU in the DFUC2020 challenge dataset, which consists of 4,500 DFU images. We further refined the EfficientDet architecture to avoid false negative and false positive predictions. The code for this method is available at https://github.com/Manugoyal12345/Yet-Another-EfficientDet-Pytorch.
Artificial Intelligence-Based Image Classification for Diagnosis of Skin Cancer: Challenges and Opportunities
Dec 05, 2019



Abstract:Recently, there has been great interest in developing Artificial Intelligence (AI) enabled computer-aided diagnostics solutions for the diagnosis of skin cancer. With the increasing incidence of skin cancers, low awareness among a growing population, and a lack of adequate clinical expertise and services, there is an immediate need for AI systems to assist clinicians in this domain. A large number of skin lesion datasets are available publicly, and researchers have developed AI-based image classification solutions, particularly deep learning algorithms, to distinguish malignant skin lesions from benign lesions in different image modalities such as dermoscopic, clinical, and histopathology images. Despite the various claims of AI systems achieving higher accuracy than dermatologists in the classification of different skin lesions, these AI systems are still in the very early stages of clinical application in terms of being ready to aid clinicians in the diagnosis of skin cancers. In this review, we discuss advancements in the digital image-based AI solutions for the diagnosis of skin cancer, along with some challenges and future opportunities to improve these AI systems to support dermatologists and enhance their ability to diagnose skin cancer.
Recognition of Ischaemia and Infection in Diabetic Foot Ulcers: Dataset and Techniques
Sep 11, 2019
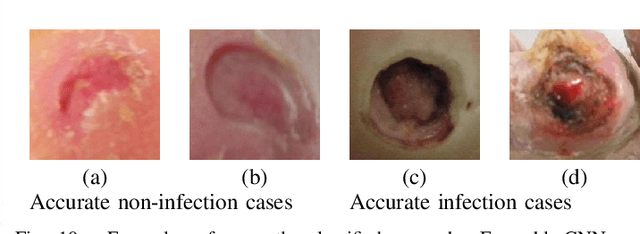
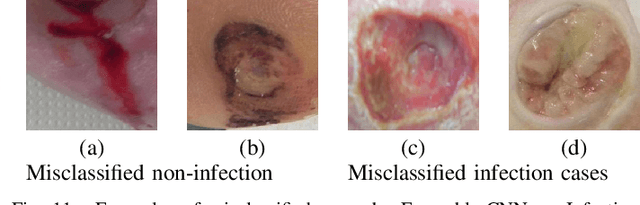
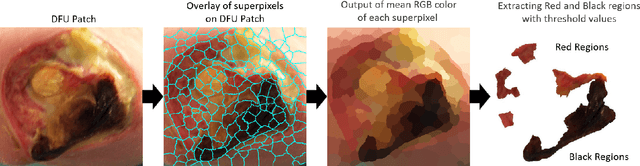
Abstract:Diabetic Foot Ulcers (DFU) detection using computerized methods is an emerging research area with the evolution of machine learning algorithms. However, existing research focuses on detecting and segmenting the ulcers. According to DFU medical classification systems, i.e. University of Texas Classification and SINBAD Classification, the presence of infection (bacteria in the wound) and ischaemia (inadequate blood supply) has important clinical implication for DFU assessment, which were used to predict the risk of amputation. In this work, we propose a new dataset and novel techniques to identify the presence of infection and ischaemia. We introduce a very comprehensive DFU dataset with ground truth labels of ischaemia and infection cases. For hand-crafted machine learning approach, we propose new feature descriptor, namely Superpixel Color Descriptor. Then, we propose a technique using Ensemble Convolutional Neural Network (CNN) model for ischaemia and infection recognition. The novelty lies in our proposed natural data-augmentation method, which clearly identifies the region of interest on foot images and focuses on finding the salient features existing in this area. Finally, we evaluate the performance of our proposed techniques on binary classification, i.e. ischaemia versus non-ischaemia and infection versus non-infection. Overall, our proposed method performs better in the classification of ischaemia than infection. We found that our proposed Ensemble CNN deep learning algorithms performed better for both classification tasks than hand-crafted machine learning algorithms, with 90% accuracy in ischaemia classification and 73% in infection classification.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge