Dmitriy A. Yablonskiy
DiffGEPCI: 3D MRI Synthesis from mGRE Signals using 2.5D Diffusion Model
Nov 29, 2023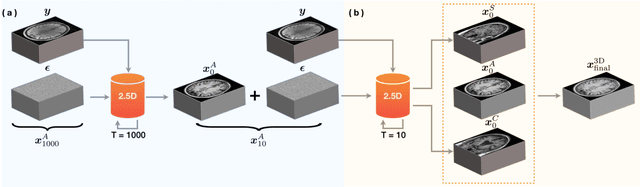
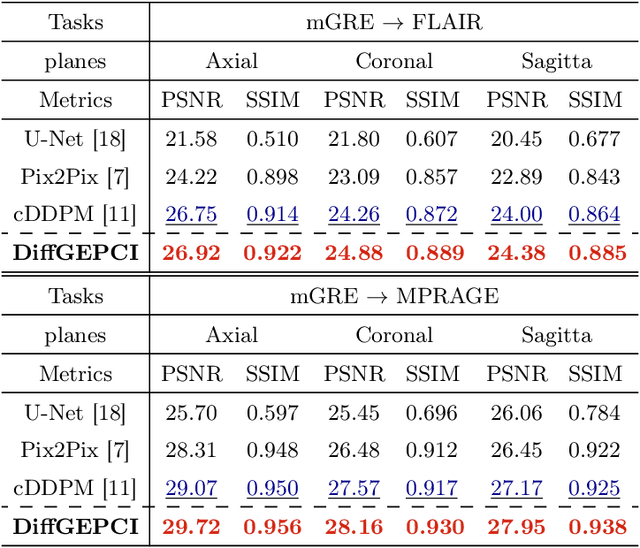
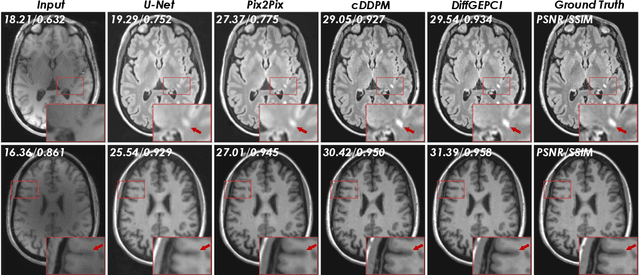
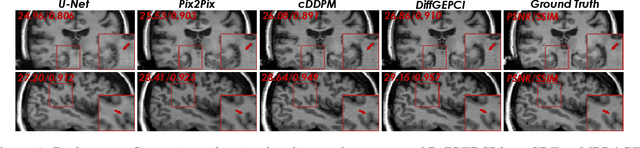
Abstract:We introduce a new framework called DiffGEPCI for cross-modality generation in magnetic resonance imaging (MRI) using a 2.5D conditional diffusion model. DiffGEPCI can synthesize high-quality Fluid Attenuated Inversion Recovery (FLAIR) and Magnetization Prepared-Rapid Gradient Echo (MPRAGE) images, without acquiring corresponding measurements, by leveraging multi-Gradient-Recalled Echo (mGRE) MRI signals as conditional inputs. DiffGEPCI operates in a two-step fashion: it initially estimates a 3D volume slice-by-slice using the axial plane and subsequently applies a refinement algorithm (referred to as 2.5D) to enhance the quality of the coronal and sagittal planes. Experimental validation on real mGRE data shows that DiffGEPCI achieves excellent performance, surpassing generative adversarial networks (GANs) and traditional diffusion models.
CoRRECT: A Deep Unfolding Framework for Motion-Corrected Quantitative R2* Mapping
Oct 12, 2022



Abstract:Quantitative MRI (qMRI) refers to a class of MRI methods for quantifying the spatial distribution of biological tissue parameters. Traditional qMRI methods usually deal separately with artifacts arising from accelerated data acquisition, involuntary physical motion, and magnetic-field inhomogeneities, leading to suboptimal end-to-end performance. This paper presents CoRRECT, a unified deep unfolding (DU) framework for qMRI consisting of a model-based end-to-end neural network, a method for motion-artifact reduction, and a self-supervised learning scheme. The network is trained to produce R2* maps whose k-space data matches the real data by also accounting for motion and field inhomogeneities. When deployed, CoRRECT only uses the k-space data without any pre-computed parameters for motion or inhomogeneity correction. Our results on experimentally collected multi-Gradient-Recalled Echo (mGRE) MRI data show that CoRRECT recovers motion and inhomogeneity artifact-free R2* maps in highly accelerated acquisition settings. This work opens the door to DU methods that can integrate physical measurement models, biophysical signal models, and learned prior models for high-quality qMRI.
Learning-based Motion Artifact Removal Networks (LEARN) for Quantitative $R_2^\ast$ Mapping
Sep 03, 2021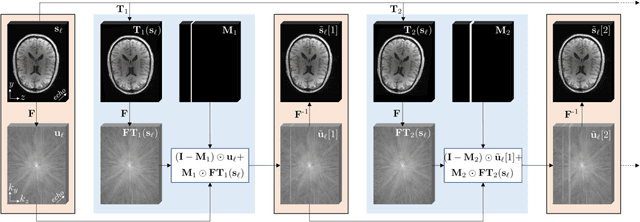
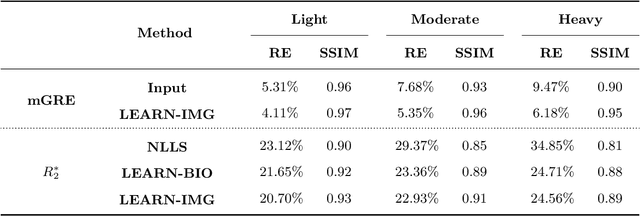
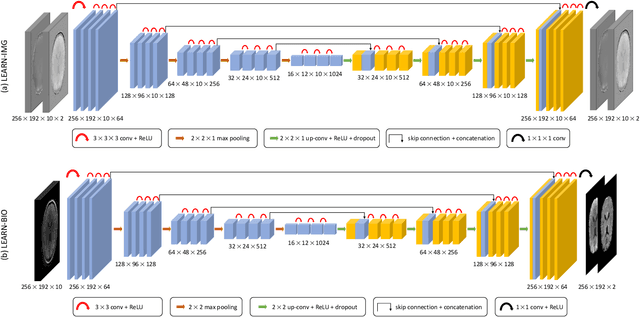
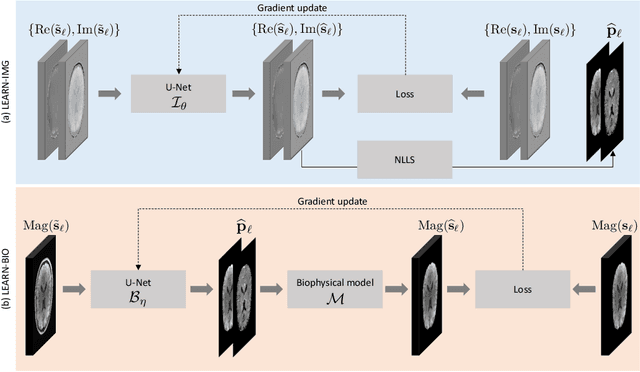
Abstract:Purpose: To introduce two novel learning-based motion artifact removal networks (LEARN) for the estimation of quantitative motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ maps from motion-corrupted multi-Gradient-Recalled Echo (mGRE) MRI data. Methods: We train two convolutional neural networks (CNNs) to correct motion artifacts for high-quality estimation of quantitative $B0$-inhomogeneity-corrected $R_2^\ast$ maps from mGRE sequences. The first CNN, LEARN-IMG, performs motion correction on complex mGRE images, to enable the subsequent computation of high-quality motion-free quantitative $R_2^\ast$ (and any other mGRE-enabled) maps using the standard voxel-wise analysis or machine-learning-based analysis. The second CNN, LEARN-BIO, is trained to directly generate motion- and $B0$-inhomogeneity-corrected quantitative $R_2^\ast$ maps from motion-corrupted magnitude-only mGRE images by taking advantage of the biophysical model describing the mGRE signal decay. We show that both CNNs trained on synthetic MR images are capable of suppressing motion artifacts while preserving details in the predicted quantitative $R_2^\ast$ maps. Significant reduction of motion artifacts on experimental in vivo motion-corrupted data has also been achieved by using our trained models. Conclusion: Both LEARN-IMG and LEARN-BIO can enable the computation of high-quality motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ maps. LEARN-IMG performs motion correction on mGRE images and relies on the subsequent analysis for the estimation of $R_2^\ast$ maps, while LEARN-BIO directly performs motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ estimation. Both LEARN-IMG and LEARN-BIO jointly process all the available gradient echoes, which enables them to exploit spatial patterns available in the data. The high computational speed of LEARN-BIO is an advantage that can lead to a broader clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge