Sayan Kahali
Learning-based Motion Artifact Removal Networks (LEARN) for Quantitative $R_2^\ast$ Mapping
Sep 03, 2021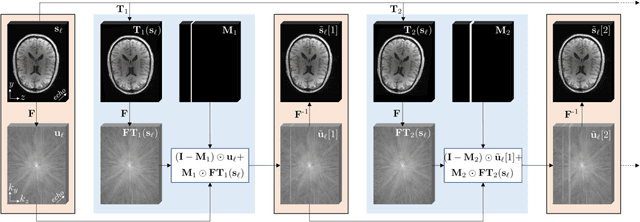
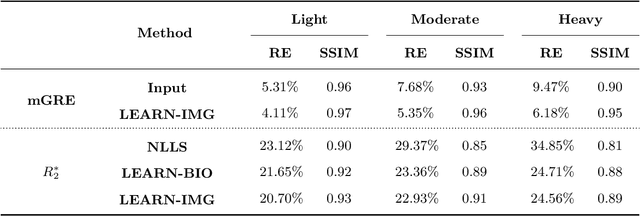
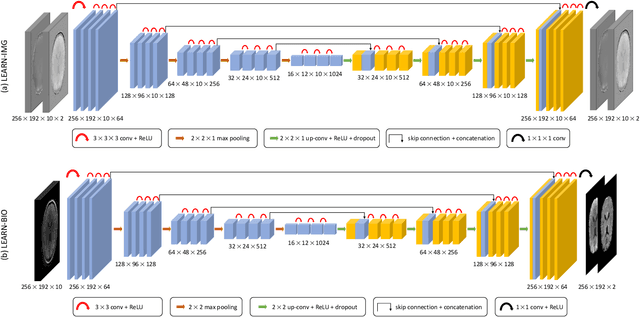
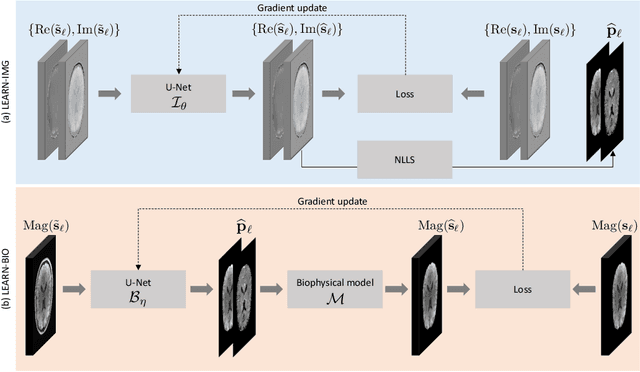
Abstract:Purpose: To introduce two novel learning-based motion artifact removal networks (LEARN) for the estimation of quantitative motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ maps from motion-corrupted multi-Gradient-Recalled Echo (mGRE) MRI data. Methods: We train two convolutional neural networks (CNNs) to correct motion artifacts for high-quality estimation of quantitative $B0$-inhomogeneity-corrected $R_2^\ast$ maps from mGRE sequences. The first CNN, LEARN-IMG, performs motion correction on complex mGRE images, to enable the subsequent computation of high-quality motion-free quantitative $R_2^\ast$ (and any other mGRE-enabled) maps using the standard voxel-wise analysis or machine-learning-based analysis. The second CNN, LEARN-BIO, is trained to directly generate motion- and $B0$-inhomogeneity-corrected quantitative $R_2^\ast$ maps from motion-corrupted magnitude-only mGRE images by taking advantage of the biophysical model describing the mGRE signal decay. We show that both CNNs trained on synthetic MR images are capable of suppressing motion artifacts while preserving details in the predicted quantitative $R_2^\ast$ maps. Significant reduction of motion artifacts on experimental in vivo motion-corrupted data has also been achieved by using our trained models. Conclusion: Both LEARN-IMG and LEARN-BIO can enable the computation of high-quality motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ maps. LEARN-IMG performs motion correction on mGRE images and relies on the subsequent analysis for the estimation of $R_2^\ast$ maps, while LEARN-BIO directly performs motion- and $B0$-inhomogeneity-corrected $R_2^\ast$ estimation. Both LEARN-IMG and LEARN-BIO jointly process all the available gradient echoes, which enables them to exploit spatial patterns available in the data. The high computational speed of LEARN-BIO is an advantage that can lead to a broader clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge