Joseph DiPalma
Improving Representation Learning for Histopathologic Images with Cluster Constraints
Oct 18, 2023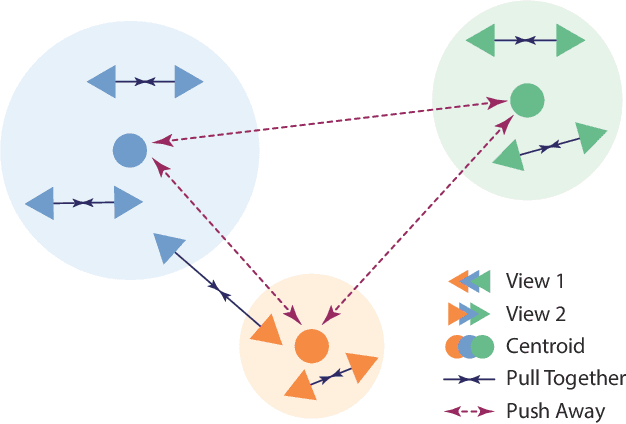
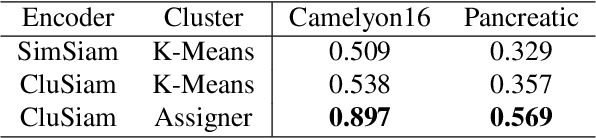
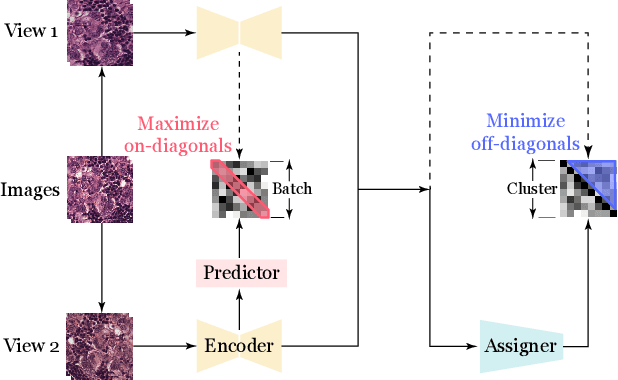
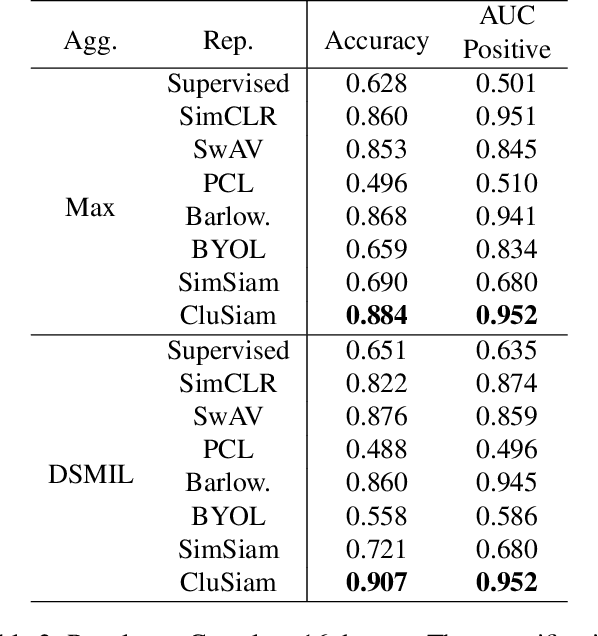
Abstract:Recent advances in whole-slide image (WSI) scanners and computational capabilities have significantly propelled the application of artificial intelligence in histopathology slide analysis. While these strides are promising, current supervised learning approaches for WSI analysis come with the challenge of exhaustively labeling high-resolution slides - a process that is both labor-intensive and time-consuming. In contrast, self-supervised learning (SSL) pretraining strategies are emerging as a viable alternative, given that they don't rely on explicit data annotations. These SSL strategies are quickly bridging the performance disparity with their supervised counterparts. In this context, we introduce an SSL framework. This framework aims for transferable representation learning and semantically meaningful clustering by synergizing invariance loss and clustering loss in WSI analysis. Notably, our approach outperforms common SSL methods in downstream classification and clustering tasks, as evidenced by tests on the Camelyon16 and a pancreatic cancer dataset. The code and additional details are accessible at: https://github.com/wwyi1828/CluSiam.
* Accepted by ICCV2023
HistoPerm: A Permutation-Based View Generation Approach for Learning Histopathologic Feature Representations
Sep 13, 2022



Abstract:Recently, deep learning methods have been successfully applied to solve numerous challenges in the field of digital pathology. However, many of these approaches are fully supervised and require annotated images. Annotating a histology image is a time-consuming and tedious process for even a highly skilled pathologist, and, as such, most histology datasets lack region-of-interest annotations and are weakly labeled. In this paper, we introduce HistoPerm, a view generation approach designed for improving the performance of representation learning techniques on histology images in weakly supervised settings. In HistoPerm, we permute augmented views of patches generated from whole-slide histology images to improve classification accuracy. These permuted views belong to the same original slide-level class but are produced from distinct patch instances. We tested adding HistoPerm to BYOL and SimCLR, two prominent representation learning methods, on two public histology datasets for Celiac disease and Renal Cell Carcinoma. For both datasets, we found improved performance in terms of accuracy, F1-score, and AUC compared to the standard BYOL and SimCLR approaches. Particularly, in a linear evaluation configuration, HistoPerm increases classification accuracy on the Celiac disease dataset by 8% for BYOL and 3% for SimCLR. Similarly, with HistoPerm, classification accuracy increases by 2% for BYOL and 0.25% for SimCLR on the Renal Cell Carcinoma dataset. The proposed permutation-based view generation approach can be adopted in common representation learning frameworks to capture histopathology features in weakly supervised settings and can lead to whole-slide classification outcomes that are close to, or even better than, fully supervised methods.
Resolution-Based Distillation for Efficient Histology Image Classification
Jan 11, 2021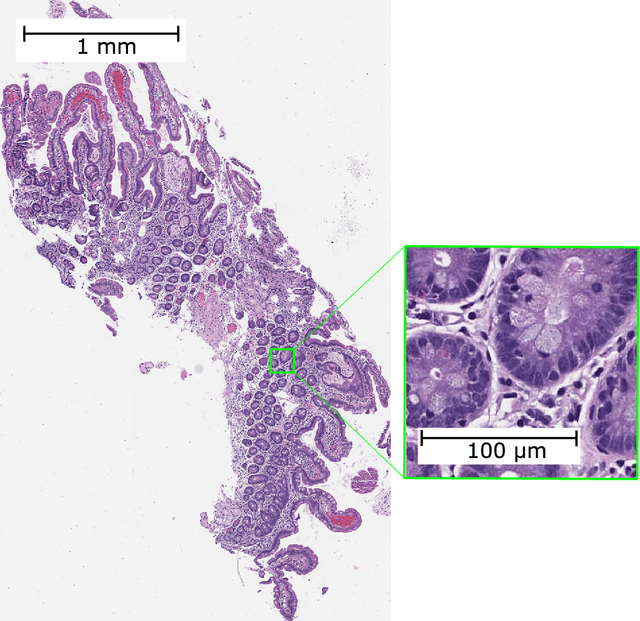
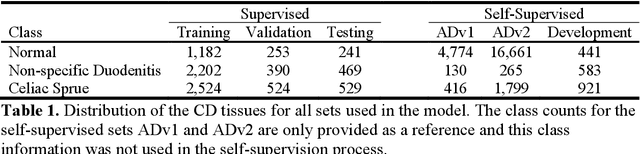
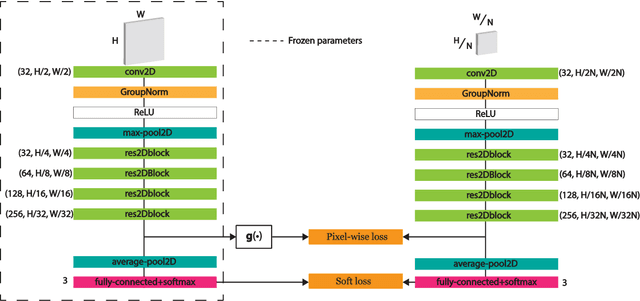
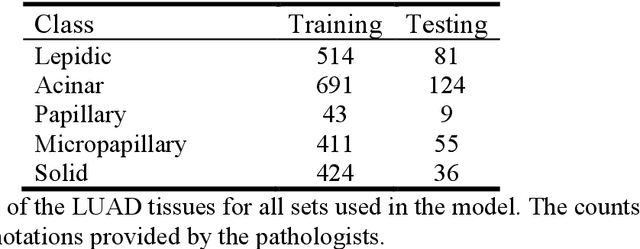
Abstract:Developing deep learning models to analyze histology images has been computationally challenging, as the massive size of the images causes excessive strain on all parts of the computing pipeline. This paper proposes a novel deep learning-based methodology for improving the computational efficiency of histology image classification. The proposed approach is robust when used with images that have reduced input resolution and can be trained effectively with limited labeled data. Pre-trained on the original high-resolution (HR) images, our method uses knowledge distillation (KD) to transfer learned knowledge from a teacher model to a student model trained on the same images at a much lower resolution. To address the lack of large-scale labeled histology image datasets, we perform KD in a self-supervised manner. We evaluate our approach on two histology image datasets associated with celiac disease (CD) and lung adenocarcinoma (LUAD). Our results show that a combination of KD and self-supervision allows the student model to approach, and in some cases, surpass the classification accuracy of the teacher, while being much more efficient. Additionally, we observe an increase in student classification performance as the size of the unlabeled dataset increases, indicating that there is potential to scale further. For the CD data, our model outperforms the HR teacher model, while needing 4 times fewer computations. For the LUAD data, our student model results at 1.25x magnification are within 3% of the teacher model at 10x magnification, with a 64 times computational cost reduction. Moreover, our CD outcomes benefit from performance scaling with the use of more unlabeled data. For 0.625x magnification, using unlabeled data improves accuracy by 4% over the baseline. Thus, our method can improve the feasibility of deep learning solutions for digital pathology with standard computational hardware.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge