Mengdan Zhu
Cross-modal RAG: Sub-dimensional Retrieval-Augmented Text-to-Image Generation
May 29, 2025Abstract:Text-to-image generation increasingly demands access to domain-specific, fine-grained, and rapidly evolving knowledge that pretrained models cannot fully capture. Existing Retrieval-Augmented Generation (RAG) methods attempt to address this by retrieving globally relevant images, but they fail when no single image contains all desired elements from a complex user query. We propose Cross-modal RAG, a novel framework that decomposes both queries and images into sub-dimensional components, enabling subquery-aware retrieval and generation. Our method introduces a hybrid retrieval strategy - combining a sub-dimensional sparse retriever with a dense retriever - to identify a Pareto-optimal set of images, each contributing complementary aspects of the query. During generation, a multimodal large language model is guided to selectively condition on relevant visual features aligned to specific subqueries, ensuring subquery-aware image synthesis. Extensive experiments on MS-COCO, Flickr30K, WikiArt, CUB, and ImageNet-LT demonstrate that Cross-modal RAG significantly outperforms existing baselines in both retrieval and generation quality, while maintaining high efficiency.
LatentExplainer: Explaining Latent Representations in Deep Generative Models with Multi-modal Foundation Models
Jun 24, 2024Abstract:Deep generative models like VAEs and diffusion models have advanced various generation tasks by leveraging latent variables to learn data distributions and generate high-quality samples. Despite the field of explainable AI making strides in interpreting machine learning models, understanding latent variables in generative models remains challenging. This paper introduces LatentExplainer, a framework for automatically generating semantically meaningful explanations of latent variables in deep generative models. LatentExplainer tackles three main challenges: inferring the meaning of latent variables, aligning explanations with inductive biases, and handling varying degrees of explainability. By perturbing latent variables and interpreting changes in generated data, the framework provides a systematic approach to understanding and controlling the data generation process, enhancing the transparency and interpretability of deep generative models. We evaluate our proposed method on several real-world and synthetic datasets, and the results demonstrate superior performance in generating high-quality explanations of latent variables.
DUE: Dynamic Uncertainty-Aware Explanation Supervision via 3D Imputation
Mar 16, 2024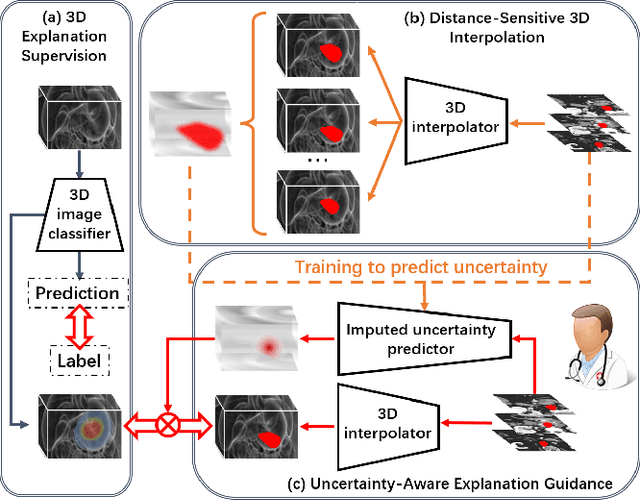
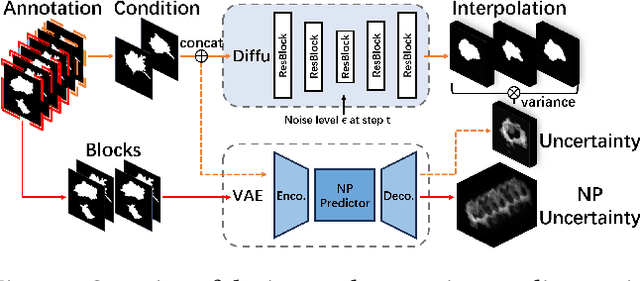
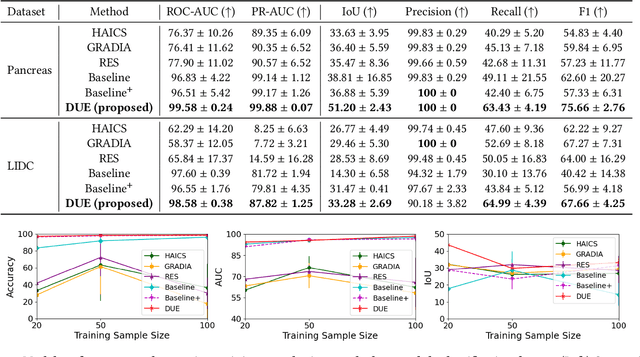
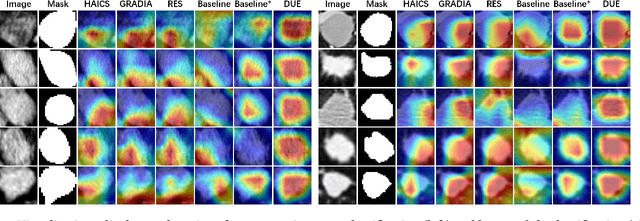
Abstract:Explanation supervision aims to enhance deep learning models by integrating additional signals to guide the generation of model explanations, showcasing notable improvements in both the predictability and explainability of the model. However, the application of explanation supervision to higher-dimensional data, such as 3D medical images, remains an under-explored domain. Challenges associated with supervising visual explanations in the presence of an additional dimension include: 1) spatial correlation changed, 2) lack of direct 3D annotations, and 3) uncertainty varies across different parts of the explanation. To address these challenges, we propose a Dynamic Uncertainty-aware Explanation supervision (DUE) framework for 3D explanation supervision that ensures uncertainty-aware explanation guidance when dealing with sparsely annotated 3D data with diffusion-based 3D interpolation. Our proposed framework is validated through comprehensive experiments on diverse real-world medical imaging datasets. The results demonstrate the effectiveness of our framework in enhancing the predictability and explainability of deep learning models in the context of medical imaging diagnosis applications.
Explaining latent representations of generative models with large multimodal models
Feb 02, 2024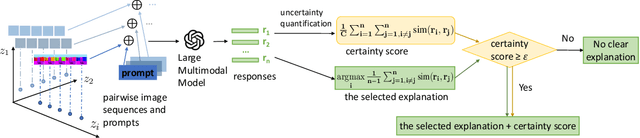
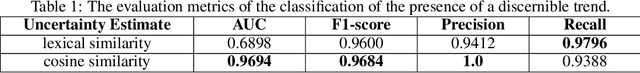
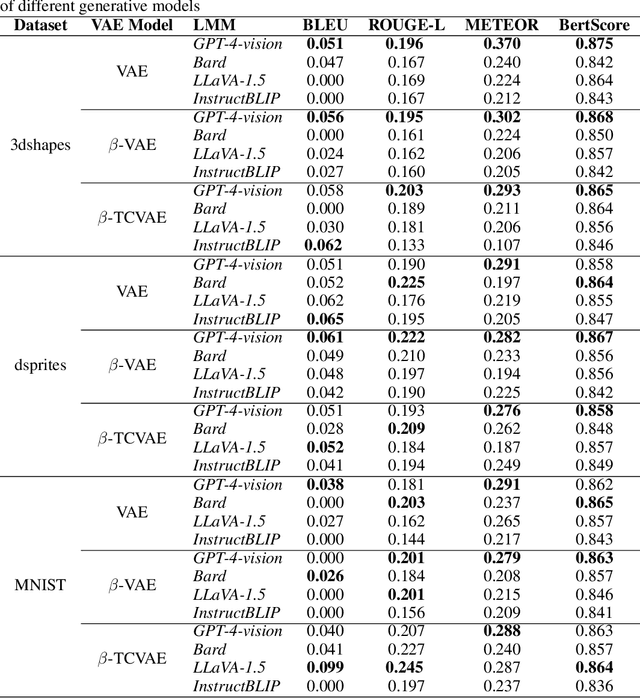

Abstract:Learning interpretable representations of data generative latent factors is an important topic for the development of artificial intelligence. With the rise of the large multimodal model, it can align images with text to generate answers. In this work, we propose a framework to comprehensively explain each latent factor in the generative models using a large multimodal model. We further measure the uncertainty of our generated explanations, quantitatively evaluate the performance of explanation generation among multiple large multimodal models, and qualitatively visualize the variations of each latent factor to learn the disentanglement effects of different generative models on explanations. Finally, we discuss the explanatory capabilities and limitations of state-of-the-art large multimodal models.
Beyond Efficiency: A Systematic Survey of Resource-Efficient Large Language Models
Jan 04, 2024Abstract:The burgeoning field of Large Language Models (LLMs), exemplified by sophisticated models like OpenAI's ChatGPT, represents a significant advancement in artificial intelligence. These models, however, bring forth substantial challenges in the high consumption of computational, memory, energy, and financial resources, especially in environments with limited resource capabilities. This survey aims to systematically address these challenges by reviewing a broad spectrum of techniques designed to enhance the resource efficiency of LLMs. We categorize methods based on their optimization focus: computational, memory, energy, financial, and network resources and their applicability across various stages of an LLM's lifecycle, including architecture design, pretraining, finetuning, and system design. Additionally, the survey introduces a nuanced categorization of resource efficiency techniques by their specific resource types, which uncovers the intricate relationships and mappings between various resources and corresponding optimization techniques. A standardized set of evaluation metrics and datasets is also presented to facilitate consistent and fair comparisons across different models and techniques. By offering a comprehensive overview of the current sota and identifying open research avenues, this survey serves as a foundational reference for researchers and practitioners, aiding them in developing more sustainable and efficient LLMs in a rapidly evolving landscape.
Development and Evaluation of a Deep Neural Network for Histologic Classification of Renal Cell Carcinoma on Biopsy and Surgical Resection Slides
Oct 30, 2020



Abstract:Renal cell carcinoma (RCC) is the most common renal cancer in adults. The histopathologic classification of RCC is essential for diagnosis, prognosis, and management of patients. Reorganization and classification of complex histologic patterns of RCC on biopsy and surgical resection slides under a microscope remains a heavily specialized, error-prone, and time-consuming task for pathologists. In this study, we developed a deep neural network model that can accurately classify digitized surgical resection slides and biopsy slides into five related classes: clear cell RCC, papillary RCC, chromophobe RCC, renal oncocytoma, and normal. In addition to the whole-slide classification pipeline, we visualized the identified indicative regions and features on slides for classification by reprocessing patch-level classification results to ensure the explainability of our diagnostic model. We evaluated our model on independent test sets of 78 surgical resection whole slides and 79 biopsy slides from our tertiary medical institution, and 69 randomly selected surgical resection slides from The Cancer Genome Atlas (TCGA) database. The average area under the curve (AUC) of our classifier on the internal resection slides, internal biopsy slides, and external TCGA slides is 0.98, 0.98 and 0.99, respectively. Our results suggest that the high generalizability of our approach across different data sources and specimen types. More importantly, our model has the potential to assist pathologists by (1) automatically pre-screening slides to reduce false-negative cases, (2) highlighting regions of importance on digitized slides to accelerate diagnosis, and (3) providing objective and accurate diagnosis as the second opinion.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge