Myriam Bontonou
IMT Atlantique - ELEC, MILA
Explaining models relating objects and privacy
May 02, 2024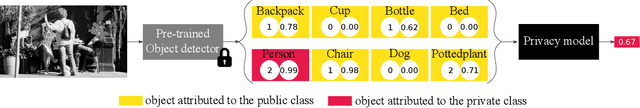
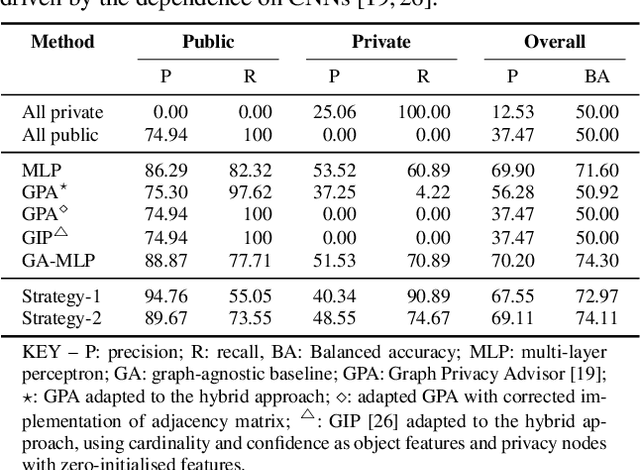
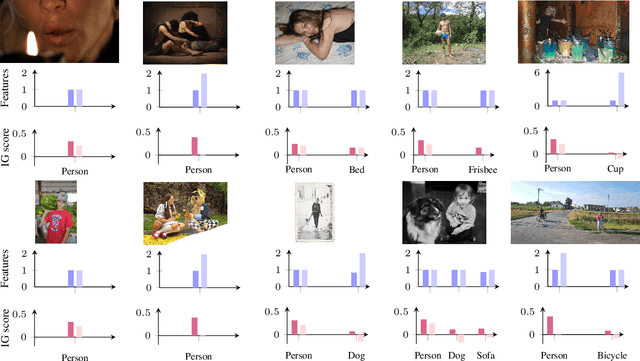
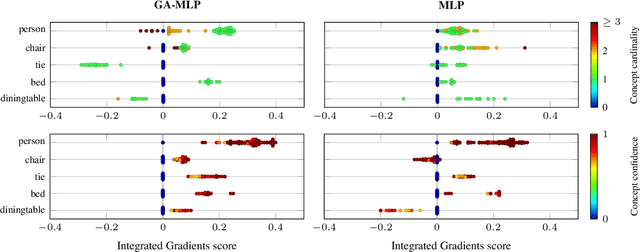
Abstract:Accurately predicting whether an image is private before sharing it online is difficult due to the vast variety of content and the subjective nature of privacy itself. In this paper, we evaluate privacy models that use objects extracted from an image to determine why the image is predicted as private. To explain the decision of these models, we use feature-attribution to identify and quantify which objects (and which of their features) are more relevant to privacy classification with respect to a reference input (i.e., no objects localised in an image) predicted as public. We show that the presence of the person category and its cardinality is the main factor for the privacy decision. Therefore, these models mostly fail to identify private images depicting documents with sensitive data, vehicle ownership, and internet activity, or public images with people (e.g., an outdoor concert or people walking in a public space next to a famous landmark). As baselines for future benchmarks, we also devise two strategies that are based on the person presence and cardinality and achieve comparable classification performance of the privacy models.
A Comparative Analysis of Gene Expression Profiling by Statistical and Machine Learning Approaches
Feb 01, 2024



Abstract:Many machine learning models have been proposed to classify phenotypes from gene expression data. In addition to their good performance, these models can potentially provide some understanding of phenotypes by extracting explanations for their decisions. These explanations often take the form of a list of genes ranked in order of importance for the predictions, the highest-ranked genes being interpreted as linked to the phenotype. We discuss the biological and the methodological limitations of such explanations. Experiments are performed on several datasets gathering cancer and healthy tissue samples from the TCGA, GTEx and TARGET databases. A collection of machine learning models including logistic regression, multilayer perceptron, and graph neural network are trained to classify samples according to their cancer type. Gene rankings are obtained from explainability methods adapted to these models, and compared to the ones from classical statistical feature selection methods such as mutual information, DESeq2, and EdgeR. Interestingly, on simple tasks, we observe that the information learned by black-box neural networks is related to the notion of differential expression. In all cases, a small set containing the best-ranked genes is sufficient to achieve a good classification. However, these genes differ significantly between the methods and similar classification performance can be achieved with numerous lower ranked genes. In conclusion, although these methods enable the identification of biomarkers characteristic of certain pathologies, our results question the completeness of the selected gene sets and thus of explainability by the identification of the underlying biological processes.
Studying Limits of Explainability by Integrated Gradients for Gene Expression Models
Mar 19, 2023


Abstract:Understanding the molecular processes that drive cellular life is a fundamental question in biological research. Ambitious programs have gathered a number of molecular datasets on large populations. To decipher the complex cellular interactions, recent work has turned to supervised machine learning methods. The scientific questions are formulated as classical learning problems on tabular data or on graphs, e.g. phenotype prediction from gene expression data. In these works, the input features on which the individual predictions are predominantly based are often interpreted as indicative of the cause of the phenotype, such as cancer identification. Here, we propose to explore the relevance of the biomarkers identified by Integrated Gradients, an explainability method for feature attribution in machine learning. Through a motivating example on The Cancer Genome Atlas, we show that ranking features by importance is not enough to robustly identify biomarkers. As it is difficult to evaluate whether biomarkers reflect relevant causes without known ground truth, we simulate gene expression data by proposing a hierarchical model based on Latent Dirichlet Allocation models. We also highlight good practices for evaluating explanations for genomics data and propose a direction to derive more insights from these explanations.
Graphs as Tools to Improve Deep Learning Methods
Oct 08, 2021


Abstract:In recent years, deep neural networks (DNNs) have known an important rise in popularity. However, although they are state-of-the-art in many machine learning challenges, they still suffer from several limitations. For example, DNNs require a lot of training data, which might not be available in some practical applications. In addition, when small perturbations are added to the inputs, DNNs are prone to misclassification errors. DNNs are also viewed as black-boxes and as such their decisions are often criticized for their lack of interpretability. In this chapter, we review recent works that aim at using graphs as tools to improve deep learning methods. These graphs are defined considering a specific layer in a deep learning architecture. Their vertices represent distinct samples, and their edges depend on the similarity of the corresponding intermediate representations. These graphs can then be leveraged using various methodologies, many of which built on top of graph signal processing. This chapter is composed of four main parts: tools for visualizing intermediate layers in a DNN, denoising data representations, optimizing graph objective functions and regularizing the learning process.
Graph-LDA: Graph Structure Priors to Improve the Accuracy in Few-Shot Classification
Aug 23, 2021



Abstract:It is very common to face classification problems where the number of available labeled samples is small compared to their dimension. These conditions are likely to cause underdetermined settings, with high risk of overfitting. To improve the generalization ability of trained classifiers, common solutions include using priors about the data distribution. Among many options, data structure priors, often represented through graphs, are increasingly popular in the field. In this paper, we introduce a generic model where observed class signals are supposed to be deteriorated with two sources of noise, one independent of the underlying graph structure and isotropic, and the other colored by a known graph operator. Under this model, we derive an optimal methodology to classify such signals. Interestingly, this methodology includes a single parameter, making it particularly suitable for cases where available data is scarce. Using various real datasets, we showcase the ability of the proposed model to be implemented in real world scenarios, resulting in increased generalization accuracy compared to popular alternatives.
Ranking Deep Learning Generalization using Label Variation in Latent Geometry Graphs
Nov 25, 2020

Abstract:Measuring the generalization performance of a Deep Neural Network (DNN) without relying on a validation set is a difficult task. In this work, we propose exploiting Latent Geometry Graphs (LGGs) to represent the latent spaces of trained DNN architectures. Such graphs are obtained by connecting samples that yield similar latent representations at a given layer of the considered DNN. We then obtain a generalization score by looking at how strongly connected are samples of distinct classes in LGGs. This score allowed us to rank 3rd on the NeurIPS 2020 Predicting Generalization in Deep Learning (PGDL) competition.
Few-shot Learning for Decoding Brain Signals
Oct 23, 2020


Abstract:Few-shot learning consists in addressing data-thrifty (inductive few-shot) or label-thrifty (transductive few-shot) problems. So far, the field has been mostly driven by applications in computer vision. In this work, we are interested in stressing the ability of recently introduced few-shot methods to solve problems dealing with neuroimaging data, a promising application field. To this end, we propose a benchmark dataset and compare multiple learning paradigms, including meta-learning, as well as various backbone networks. Our experiments show that few-shot methods are able to efficiently decode brain signals using few examples, and that graph-based backbones do not outperform simple structure-agnostic solutions, such as multi-layer perceptrons.
Predicting the Accuracy of a Few-Shot Classifier
Jul 08, 2020



Abstract:In the context of few-shot learning, one cannot measure the generalization ability of a trained classifier using validation sets, due to the small number of labeled samples. In this paper, we are interested in finding alternatives to answer the question: is my classifier generalizing well to previously unseen data? We first analyze the reasons for the variability of generalization performances. We then investigate the case of using transfer-based solutions, and consider three settings: i) supervised where we only have access to a few labeled samples, ii) semi-supervised where we have access to both a few labeled samples and a set of unlabeled samples and iii) unsupervised where we only have access to unlabeled samples. For each setting, we propose reasonable measures that we empirically demonstrate to be correlated with the generalization ability of considered classifiers. We also show that these simple measures can be used to predict generalization up to a certain confidence. We conduct our experiments on standard few-shot vision datasets.
Deep geometric knowledge distillation with graphs
Nov 08, 2019



Abstract:In most cases deep learning architectures are trained disregarding the amount of operations and energy consumption. However, some applications, like embedded systems, can be resource-constrained during inference. A popular approach to reduce the size of a deep learning architecture consists in distilling knowledge from a bigger network (teacher) to a smaller one (student). Directly training the student to mimic the teacher representation can be effective, but it requires that both share the same latent space dimensions. In this work, we focus instead on relative knowledge distillation (RKD), which considers the geometry of the respective latent spaces, allowing for dimension-agnostic transfer of knowledge. Specifically we introduce a graph-based RKD method, in which graphs are used to capture the geometry of latent spaces. Using classical computer vision benchmarks, we demonstrate the ability of the proposed method to efficiently distillate knowledge from the teacher to the student, leading to better accuracy for the same budget as compared to existing RKD alternatives.
Comparing linear structure-based and data-driven latent spatial representations for sequence prediction
Aug 19, 2019


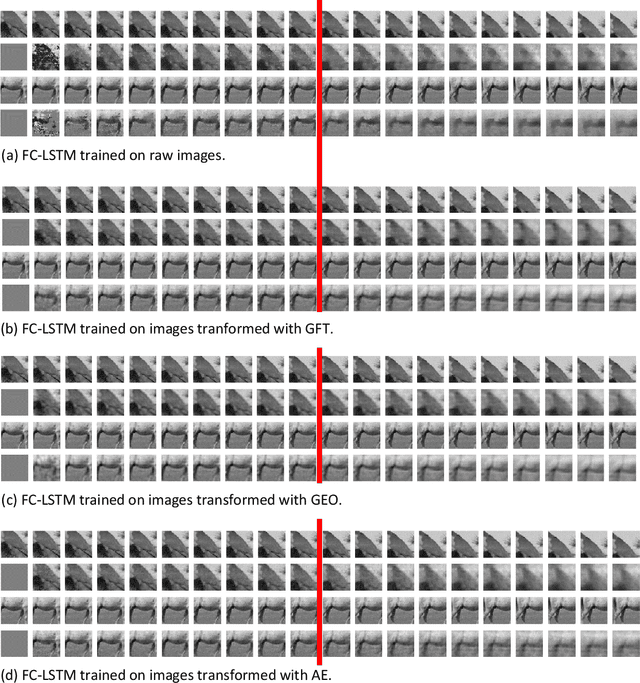
Abstract:Predicting the future of Graph-supported Time Series (GTS) is a key challenge in many domains, such as climate monitoring, finance or neuroimaging. Yet it is a highly difficult problem as it requires to account jointly for time and graph (spatial) dependencies. To simplify this process, it is common to use a two-step procedure in which spatial and time dependencies are dealt with separately. In this paper, we are interested in comparing various linear spatial representations, namely structure-based ones and data-driven ones, in terms of how they help predict the future of GTS. To that end, we perform experiments with various datasets including spontaneous brain activity and raw videos.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge