Anaïs Haget
A Comparative Analysis of Gene Expression Profiling by Statistical and Machine Learning Approaches
Feb 01, 2024



Abstract:Many machine learning models have been proposed to classify phenotypes from gene expression data. In addition to their good performance, these models can potentially provide some understanding of phenotypes by extracting explanations for their decisions. These explanations often take the form of a list of genes ranked in order of importance for the predictions, the highest-ranked genes being interpreted as linked to the phenotype. We discuss the biological and the methodological limitations of such explanations. Experiments are performed on several datasets gathering cancer and healthy tissue samples from the TCGA, GTEx and TARGET databases. A collection of machine learning models including logistic regression, multilayer perceptron, and graph neural network are trained to classify samples according to their cancer type. Gene rankings are obtained from explainability methods adapted to these models, and compared to the ones from classical statistical feature selection methods such as mutual information, DESeq2, and EdgeR. Interestingly, on simple tasks, we observe that the information learned by black-box neural networks is related to the notion of differential expression. In all cases, a small set containing the best-ranked genes is sufficient to achieve a good classification. However, these genes differ significantly between the methods and similar classification performance can be achieved with numerous lower ranked genes. In conclusion, although these methods enable the identification of biomarkers characteristic of certain pathologies, our results question the completeness of the selected gene sets and thus of explainability by the identification of the underlying biological processes.
Studying Limits of Explainability by Integrated Gradients for Gene Expression Models
Mar 19, 2023


Abstract:Understanding the molecular processes that drive cellular life is a fundamental question in biological research. Ambitious programs have gathered a number of molecular datasets on large populations. To decipher the complex cellular interactions, recent work has turned to supervised machine learning methods. The scientific questions are formulated as classical learning problems on tabular data or on graphs, e.g. phenotype prediction from gene expression data. In these works, the input features on which the individual predictions are predominantly based are often interpreted as indicative of the cause of the phenotype, such as cancer identification. Here, we propose to explore the relevance of the biomarkers identified by Integrated Gradients, an explainability method for feature attribution in machine learning. Through a motivating example on The Cancer Genome Atlas, we show that ranking features by importance is not enough to robustly identify biomarkers. As it is difficult to evaluate whether biomarkers reflect relevant causes without known ground truth, we simulate gene expression data by proposing a hierarchical model based on Latent Dirichlet Allocation models. We also highlight good practices for evaluating explanations for genomics data and propose a direction to derive more insights from these explanations.
Four-Arm Manipulation via Feet Interfaces
Sep 11, 2019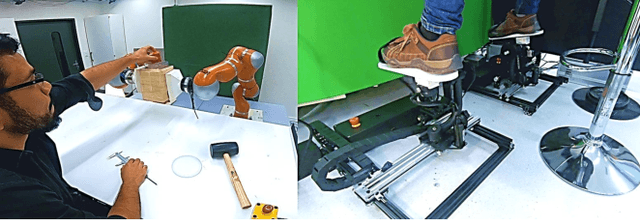
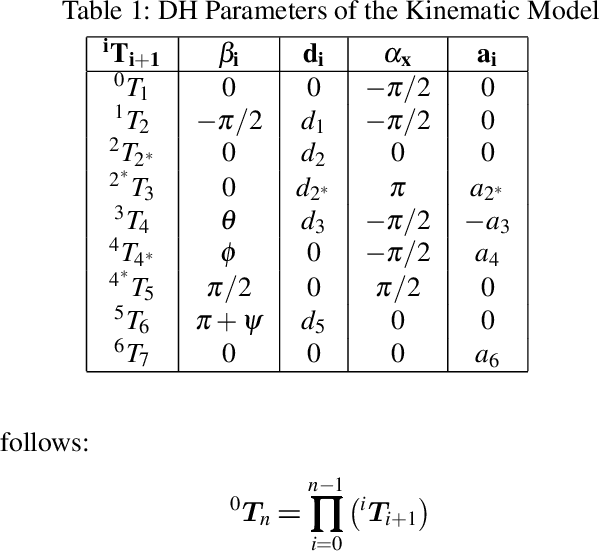

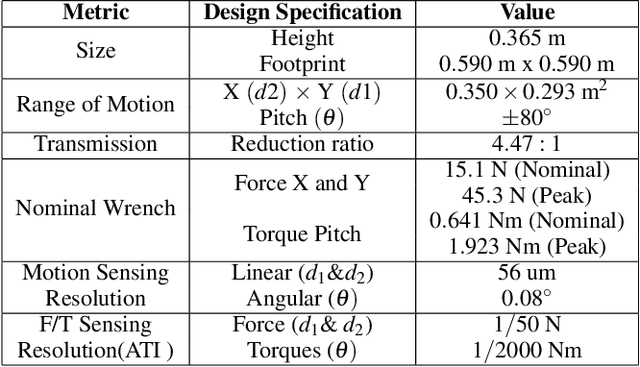
Abstract:We seek to augment human manipulation by enabling humans to control two robotic arms in addition to their natural arms using their feet. Thereby, the hands are free to perform tasks of high dexterity, while the feet-controlled arms perform tasks requiring lower dexterity, such as supporting a load. The robotic arms are tele-operated through two foot interfaces that transmit translation and rotation to the end effector of the manipulator. Haptic feedback is provided for the human to perceive contact and change in load and to adapt the feet pressure accordingly. Existing foot interfaces have been used primarily for a single foot control and are limited in range of motion and number of degrees of freedom they can control. This paper presents foot-interfaces specifically made for bipedal control, with a workspace suitable for two feet operation and in five degrees of freedom each. This paper also presents a position-force teleoperation controller based on Impedance Control modulated through Dynamical Systems for trajectory generation. Finally, an initial validation of the platform is presented, whereby a user grasps an object with both feet and generates various disturbances while the object is supported by the feet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge