Michael Widrich
Cross-Domain Few-Shot Learning by Representation Fusion
Oct 13, 2020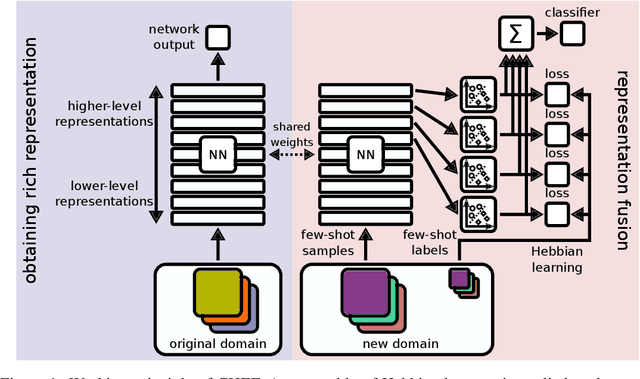

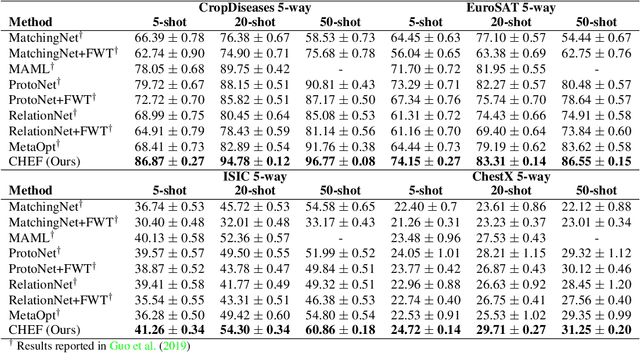
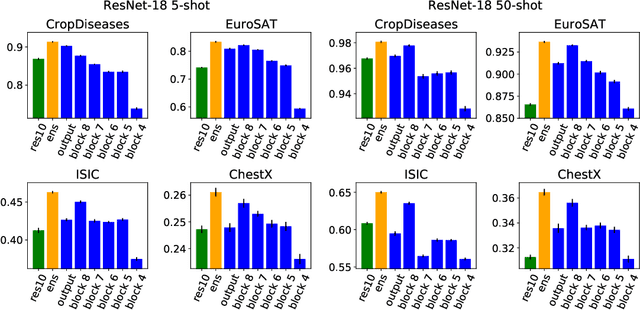
Abstract:In order to quickly adapt to new data, few-shot learning aims at learning from few examples, often by using already acquired knowledge. The new data often differs from the previously seen data due to a domain shift, that is, a change of the input-target distribution. While several methods perform well on small domain shifts like new target classes with similar inputs, larger domain shifts are still challenging. Large domain shifts may result in high-level concepts that are not shared between the original and the new domain. However, low-level concepts like edges in images might still be shared and useful. For cross-domain few-shot learning, we suggest representation fusion to unify different abstraction levels of a deep neural network into one representation. We propose Cross-domain Hebbian Ensemble Few-shot learning (CHEF), which achieves representation fusion by an ensemble of Hebbian learners acting on different layers of a deep neural network that was trained on the original domain. On the few-shot datasets miniImagenet and tieredImagenet, where the domain shift is small, CHEF is competitive with state-of-the-art methods. On cross-domain few-shot benchmark challenges with larger domain shifts, CHEF establishes novel state-of-the-art results in all categories. We further apply CHEF on a real-world cross-domain application in drug discovery. We consider a domain shift from bioactive molecules to environmental chemicals and drugs with twelve associated toxicity prediction tasks. On these tasks, that are highly relevant for computational drug discovery, CHEF significantly outperforms all its competitors. Github: https://github.com/ml-jku/chef
Modern Hopfield Networks and Attention for Immune Repertoire Classification
Jul 16, 2020



Abstract:A central mechanism in machine learning is to identify, store, and recognize patterns. How to learn, access, and retrieve such patterns is crucial in Hopfield networks and the more recent transformer architectures. We show that the attention mechanism of transformer architectures is actually the update rule of modern Hopfield networks that can store exponentially many patterns. We exploit this high storage capacity of modern Hopfield networks to solve a challenging multiple instance learning (MIL) problem in computational biology: immune repertoire classification. Accurate and interpretable machine learning methods solving this problem could pave the way towards new vaccines and therapies, which is currently a very relevant research topic intensified by the COVID-19 crisis. Immune repertoire classification based on the vast number of immunosequences of an individual is a MIL problem with an unprecedentedly massive number of instances, two orders of magnitude larger than currently considered problems, and with an extremely low witness rate. In this work, we present our novel method DeepRC that integrates transformer-like attention, or equivalently modern Hopfield networks, into deep learning architectures for massive MIL such as immune repertoire classification. We demonstrate that DeepRC outperforms all other methods with respect to predictive performance on large-scale experiments, including simulated and real-world virus infection data, and enables the extraction of sequence motifs that are connected to a given disease class. Source code and datasets: https://github.com/ml-jku/DeepRC
Hopfield Networks is All You Need
Jul 16, 2020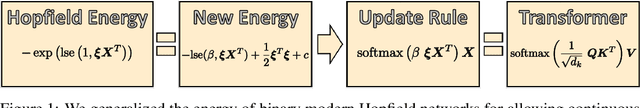
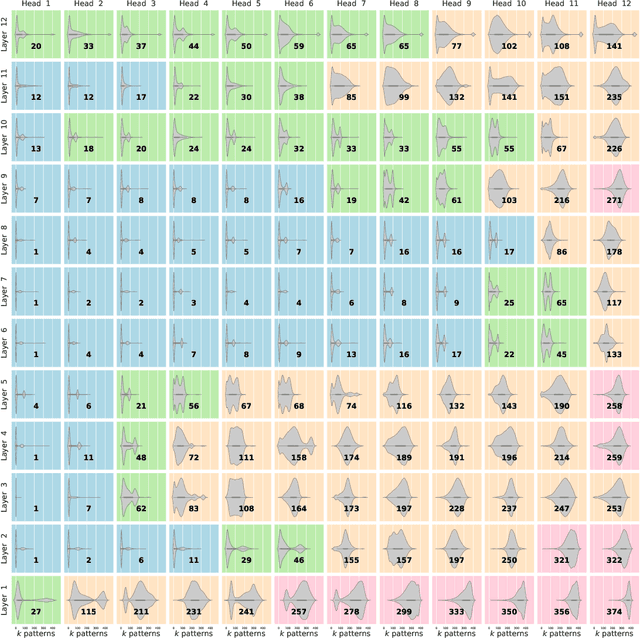
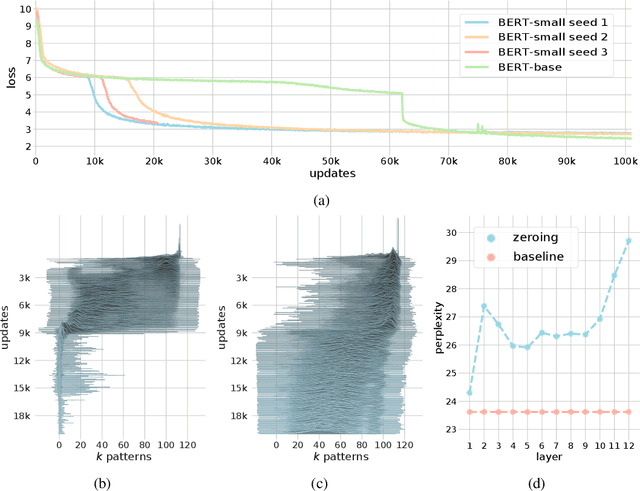
Abstract:We show that the transformer attention mechanism is the update rule of a modern Hopfield network with continuous states. This new Hopfield network can store exponentially (with the dimension) many patterns, converges with one update, and has exponentially small retrieval errors. The number of stored patterns is traded off against convergence speed and retrieval error. The new Hopfield network has three types of energy minima (fixed points of the update): (1) global fixed point averaging over all patterns, (2) metastable states averaging over a subset of patterns, and (3) fixed points which store a single pattern. Transformer and BERT models operate in their first layers preferably in the global averaging regime, while they operate in higher layers in metastable states. The gradient in transformers is maximal for metastable states, is uniformly distributed for global averaging, and vanishes for a fixed point near a stored pattern. Using the Hopfield network interpretation, we analyzed learning of transformer and BERT models. Learning starts with attention heads that average and then most of them switch to metastable states. However, the majority of heads in the first layers still averages and can be replaced by averaging, e.g. our proposed Gaussian weighting. In contrast, heads in the last layers steadily learn and seem to use metastable states to collect information created in lower layers. These heads seem to be a promising target for improving transformers. Neural networks with Hopfield networks outperform other methods on immune repertoire classification, where the Hopfield net stores several hundreds of thousands of patterns. We provide a new PyTorch layer called "Hopfield", which allows to equip deep learning architectures with modern Hopfield networks as a new powerful concept comprising pooling, memory, and attention. GitHub: https://github.com/ml-jku/hopfield-layers
Large-scale ligand-based virtual screening for SARS-CoV-2 inhibitors using deep neural networks
Apr 03, 2020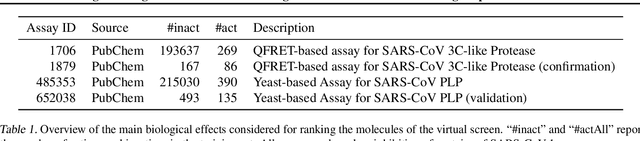
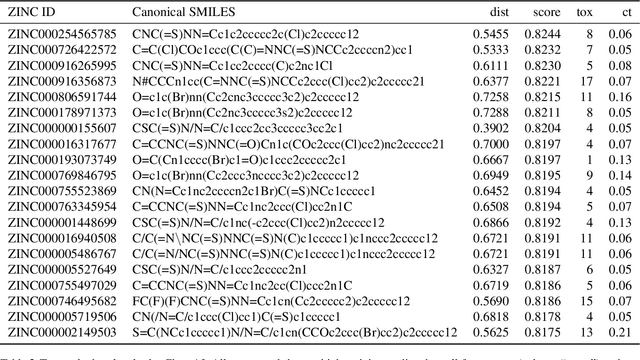
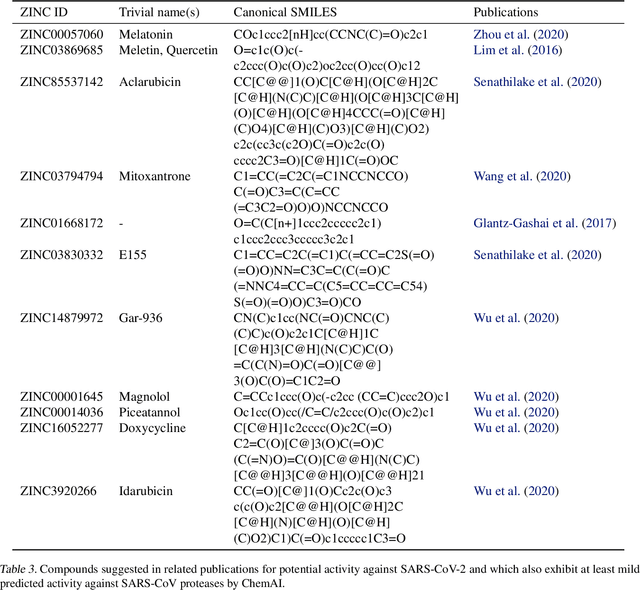
Abstract:Due to the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, there is an urgent need for novel therapies and drugs. We conducted a large-scale virtual screening for small molecules that are potential CoV-2 inhibitors. To this end, we utilized "ChemAI", a deep neural network trained on more than 220M data points across 3.6M molecules from three public drug-discovery databases. With ChemAI, we screened and ranked one billion molecules from the ZINC database for favourable effects against CoV-2. We then reduced the result to the 30,000 top-ranked compounds, which are readily accessible and purchasable via the ZINC database. Additionally, we screened the DrugBank using ChemAI to allow for drug repurposing, which would be a fast way towards a therapy. We provide these top-ranked compounds of ZINC and DrugBank as a library for further screening with bioassays at https://github.com/ml-jku/sars-cov-inhibitors-chemai.
Explaining and Interpreting LSTMs
Sep 25, 2019
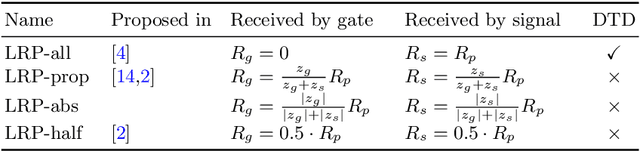
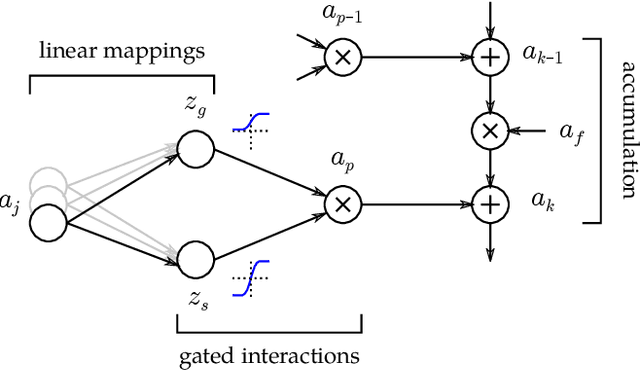
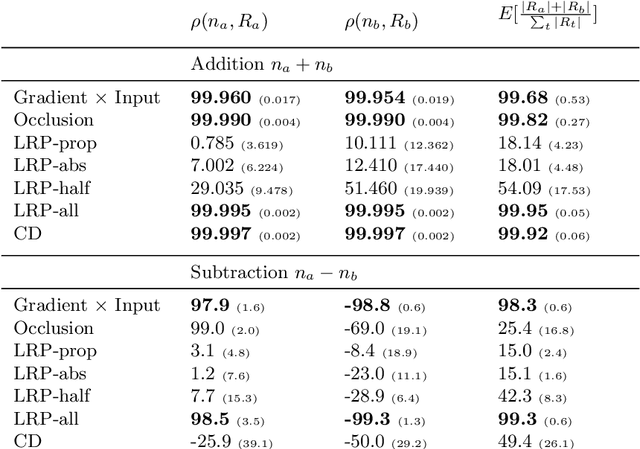
Abstract:While neural networks have acted as a strong unifying force in the design of modern AI systems, the neural network architectures themselves remain highly heterogeneous due to the variety of tasks to be solved. In this chapter, we explore how to adapt the Layer-wise Relevance Propagation (LRP) technique used for explaining the predictions of feed-forward networks to the LSTM architecture used for sequential data modeling and forecasting. The special accumulators and gated interactions present in the LSTM require both a new propagation scheme and an extension of the underlying theoretical framework to deliver faithful explanations.
RUDDER: Return Decomposition for Delayed Rewards
Jun 20, 2018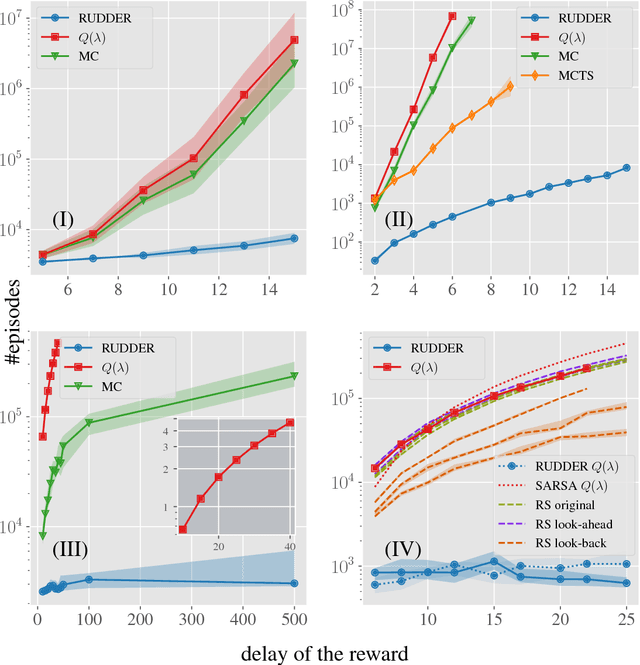

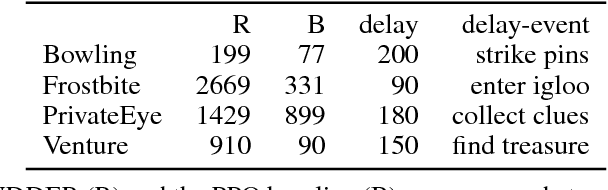
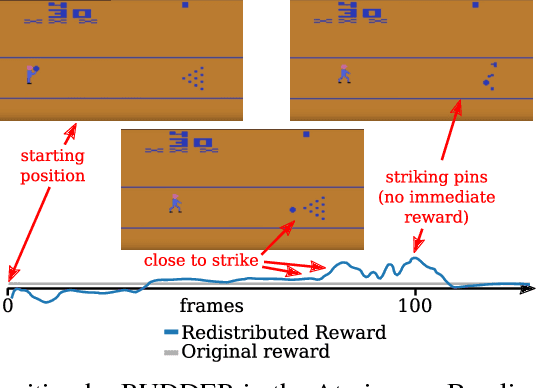
Abstract:We propose a novel reinforcement learning approach for finite Markov decision processes (MDPs) with delayed rewards. In this work, biases of temporal difference (TD) estimates are proved to be corrected only exponentially slowly in the number of delay steps. Furthermore, variances of Monte Carlo (MC) estimates are proved to increase the variance of other estimates, the number of which can exponentially grow in the number of delay steps. We introduce RUDDER, a return decomposition method, which creates a new MDP with same optimal policies as the original MDP but with redistributed rewards that have largely reduced delays. If the return decomposition is optimal, then the new MDP does not have delayed rewards and TD estimates are unbiased. In this case, the rewards track Q-values so that the future expected reward is always zero. We experimentally confirm our theoretical results on bias and variance of TD and MC estimates. On artificial tasks with different lengths of reward delays, we show that RUDDER is exponentially faster than TD, MC, and MC Tree Search (MCTS). RUDDER outperforms rainbow, A3C, DDQN, Distributional DQN, Dueling DDQN, Noisy DQN, and Prioritized DDQN on the delayed reward Atari game Venture in only a fraction of the learning time. RUDDER considerably improves the state-of-the-art on the delayed reward Atari game Bowling in much less learning time. Source code is available at https://github.com/ml-jku/baselines-rudder, with demonstration videos at https://goo.gl/EQerZV.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge