Andreu Vall
A Framework for Evaluating Predictive Models Using Synthetic Image Covariates and Longitudinal Data
Oct 21, 2024



Abstract:We present a novel framework for synthesizing patient data with complex covariates (e.g., eye scans) paired with longitudinal observations (e.g., visual acuity over time), addressing privacy concerns in healthcare research. Our approach introduces controlled association in latent spaces generating each data modality, enabling the creation of complex covariate-longitudinal observation pairs. This framework facilitates the development of predictive models and provides openly available benchmarking datasets for healthcare research. We demonstrate our framework using optical coherence tomography (OCT) scans, though it is applicable across domains. Using 109,309 2D OCT scan slices, we trained an image generative model combining a variational autoencoder and a diffusion model. Longitudinal observations were simulated using a nonlinear mixed effect (NLME) model from a low-dimensional space of random effects. We generated 1.1M OCT scan slices paired with five sets of longitudinal observations at controlled association levels (100%, 50%, 10%, 5.26%, and 2% of between-subject variability). To assess the framework, we modeled synthetic longitudinal observations with another NLME model, computed empirical Bayes estimates of random effects, and trained a ResNet to predict these estimates from synthetic OCT scans. We then incorporated ResNet predictions into the NLME model for patient-individualized predictions. Prediction accuracy on withheld data declined as intended with reduced association between images and longitudinal measurements. Notably, in all but the 2% case, we achieved within 50% of the theoretical best possible prediction on withheld data, demonstrating our ability to detect even weak signals. This confirms the effectiveness of our framework in generating synthetic data with controlled levels of association, providing a valuable tool for healthcare research.
Enhancing Activity Prediction Models in Drug Discovery with the Ability to Understand Human Language
Mar 06, 2023Abstract:Activity and property prediction models are the central workhorses in drug discovery and materials sciences, but currently they have to be trained or fine-tuned for new tasks. Without training or fine-tuning, scientific language models could be used for such low-data tasks through their announced zero- and few-shot capabilities. However, their predictive quality at activity prediction is lacking. In this work, we envision a novel type of activity prediction model that is able to adapt to new prediction tasks at inference time, via understanding textual information describing the task. To this end, we propose a new architecture with separate modules for chemical and natural language inputs, and a contrastive pre-training objective on data from large biochemical databases. In extensive experiments, we show that our method CLAMP yields improved predictive performance on few-shot learning benchmarks and zero-shot problems in drug discovery. We attribute the advances of our method to the modularized architecture and to our pre-training objective.
Large-scale ligand-based virtual screening for SARS-CoV-2 inhibitors using deep neural networks
Apr 03, 2020
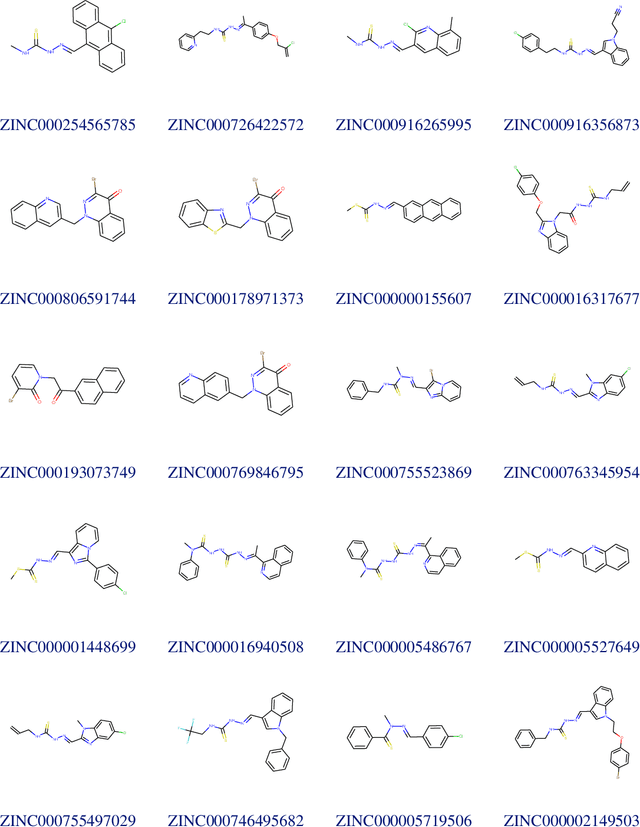
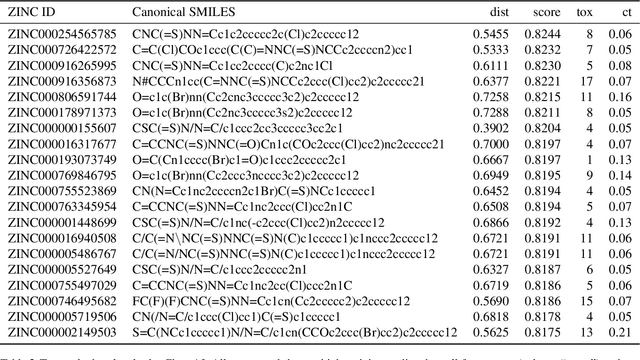
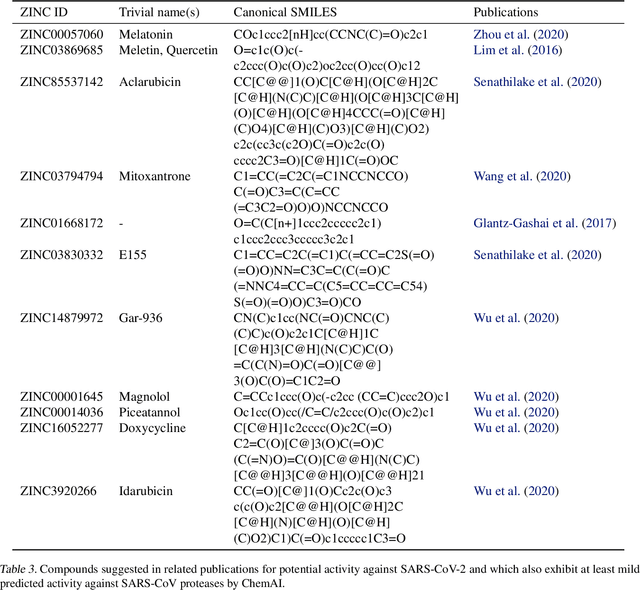
Abstract:Due to the current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic, there is an urgent need for novel therapies and drugs. We conducted a large-scale virtual screening for small molecules that are potential CoV-2 inhibitors. To this end, we utilized "ChemAI", a deep neural network trained on more than 220M data points across 3.6M molecules from three public drug-discovery databases. With ChemAI, we screened and ranked one billion molecules from the ZINC database for favourable effects against CoV-2. We then reduced the result to the 30,000 top-ranked compounds, which are readily accessible and purchasable via the ZINC database. Additionally, we screened the DrugBank using ChemAI to allow for drug repurposing, which would be a fast way towards a therapy. We provide these top-ranked compounds of ZINC and DrugBank as a library for further screening with bioassays at https://github.com/ml-jku/sars-cov-inhibitors-chemai.
Towards Explainable Music Emotion Recognition: The Route via Mid-level Features
Jul 08, 2019

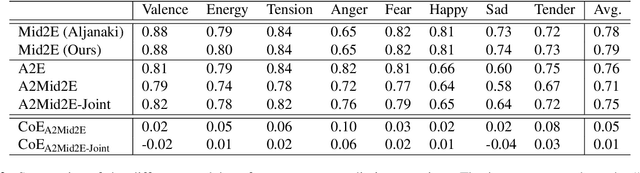
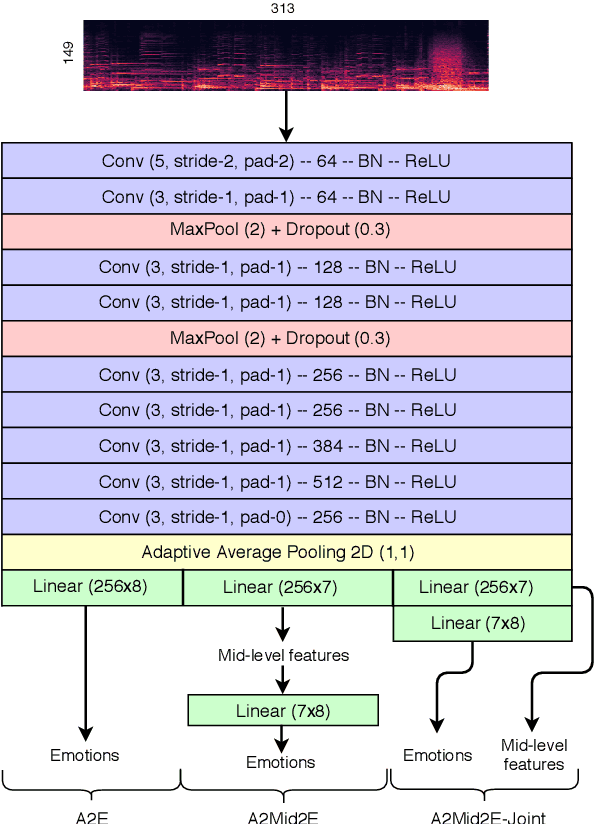
Abstract:Emotional aspects play an important part in our interaction with music. However, modelling these aspects in MIR systems have been notoriously challenging since emotion is an inherently abstract and subjective experience, thus making it difficult to quantify or predict in the first place, and to make sense of the predictions in the next. In an attempt to create a model that can give a musically meaningful and intuitive explanation for its predictions, we propose a VGG-style deep neural network that learns to predict emotional characteristics of a musical piece together with (and based on) human-interpretable, mid-level perceptual features. We compare this to predicting emotion directly with an identical network that does not take into account the mid-level features and observe that the loss in predictive performance of going through the mid-level features is surprisingly low, on average. The design of our network allows us to visualize the effects of perceptual features on individual emotion predictions, and we argue that the small loss in performance in going through the mid-level features is justified by the gain in explainability of the predictions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge