Johannes Brandstetter
WIND: Weather Inverse Diffusion for Zero-Shot Atmospheric Modeling
Feb 03, 2026Abstract:Deep learning has revolutionized weather and climate modeling, yet the current landscape remains fragmented: highly specialized models are typically trained individually for distinct tasks. To unify this landscape, we introduce WIND, a single pre-trained foundation model capable of replacing specialized baselines across a vast array of tasks. Crucially, in contrast to previous atmospheric foundation models, we achieve this without any task-specific fine-tuning. To learn a robust, task-agnostic prior of the atmosphere, we pre-train WIND with a self-supervised video reconstruction objective, utilizing an unconditional video diffusion model to iteratively reconstruct atmospheric dynamics from a noisy state. At inference, we frame diverse domain-specific problems strictly as inverse problems and solve them via posterior sampling. This unified approach allows us to tackle highly relevant weather and climate problems, including probabilistic forecasting, spatial and temporal downscaling, sparse reconstruction and enforcing conservation laws purely with our pre-trained model. We further demonstrate the model's capacity to generate physically consistent counterfactual storylines of extreme weather events under global warming scenarios. By combining generative video modeling with inverse problem solving, WIND offers a computationally efficient paradigm shift in AI-based atmospheric modeling.
Autoregressive deep learning for real-time simulation of soft tissue dynamics during virtual neurosurgery
Jan 20, 2026Abstract:Accurate simulation of brain deformation is a key component for developing realistic, interactive neurosurgical simulators, as complex nonlinear deformations must be captured to ensure realistic tool-tissue interactions. However, traditional numerical solvers often fall short in meeting real-time performance requirements. To overcome this, we introduce a deep learning-based surrogate model that efficiently simulates transient brain deformation caused by continuous interactions between surgical instruments and the virtual brain geometry. Building on Universal Physics Transformers, our approach operates directly on large-scale mesh data and is trained on an extensive dataset generated from nonlinear finite element simulations, covering a broad spectrum of temporal instrument-tissue interaction scenarios. To reduce the accumulation of errors in autoregressive inference, we propose a stochastic teacher forcing strategy applied during model training. Specifically, training consists of short stochastic rollouts in which the proportion of ground truth inputs is gradually decreased in favor of model-generated predictions. Our results show that the proposed surrogate model achieves accurate and efficient predictions across a range of transient brain deformation scenarios, scaling to meshes with up to 150,000 nodes. The introduced stochastic teacher forcing technique substantially improves long-term rollout stability, reducing the maximum prediction error from 6.7 mm to 3.5 mm. We further integrate the trained surrogate model into an interactive neurosurgical simulation environment, achieving runtimes below 10 ms per simulation step on consumer-grade inference hardware. Our proposed deep learning framework enables rapid, smooth and accurate biomechanical simulations of dynamic brain tissue deformation, laying the foundation for realistic surgical training environments.
GyroSwin: 5D Surrogates for Gyrokinetic Plasma Turbulence Simulations
Oct 08, 2025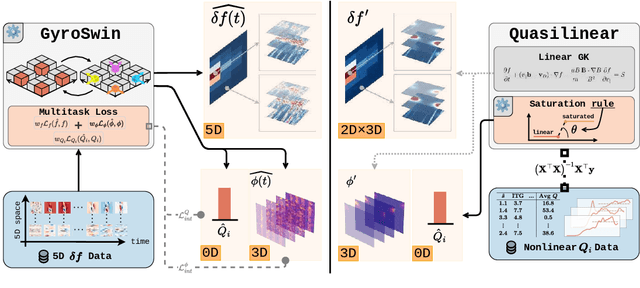
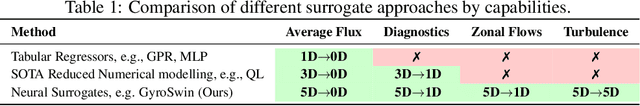
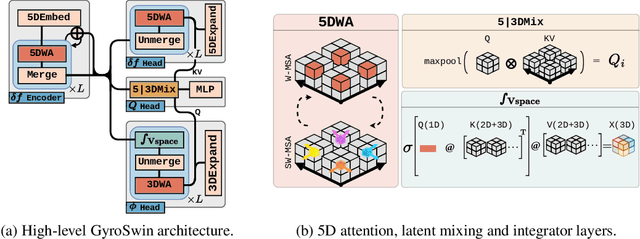
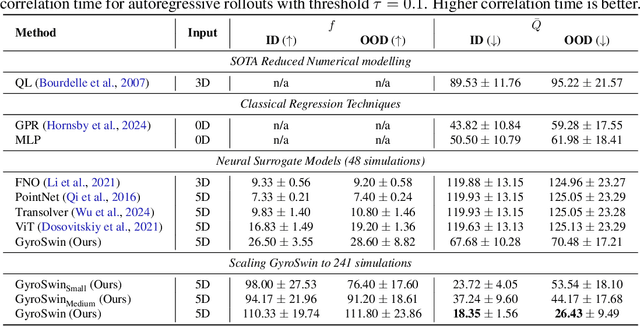
Abstract:Nuclear fusion plays a pivotal role in the quest for reliable and sustainable energy production. A major roadblock to viable fusion power is understanding plasma turbulence, which significantly impairs plasma confinement, and is vital for next-generation reactor design. Plasma turbulence is governed by the nonlinear gyrokinetic equation, which evolves a 5D distribution function over time. Due to its high computational cost, reduced-order models are often employed in practice to approximate turbulent transport of energy. However, they omit nonlinear effects unique to the full 5D dynamics. To tackle this, we introduce GyroSwin, the first scalable 5D neural surrogate that can model 5D nonlinear gyrokinetic simulations, thereby capturing the physical phenomena neglected by reduced models, while providing accurate estimates of turbulent heat transport.GyroSwin (i) extends hierarchical Vision Transformers to 5D, (ii) introduces cross-attention and integration modules for latent 3D$\leftrightarrow$5D interactions between electrostatic potential fields and the distribution function, and (iii) performs channelwise mode separation inspired by nonlinear physics. We demonstrate that GyroSwin outperforms widely used reduced numerics on heat flux prediction, captures the turbulent energy cascade, and reduces the cost of fully resolved nonlinear gyrokinetics by three orders of magnitude while remaining physically verifiable. GyroSwin shows promising scaling laws, tested up to one billion parameters, paving the way for scalable neural surrogates for gyrokinetic simulations of plasma turbulence.
Einstein Fields: A Neural Perspective To Computational General Relativity
Jul 15, 2025Abstract:We introduce Einstein Fields, a neural representation that is designed to compress computationally intensive four-dimensional numerical relativity simulations into compact implicit neural network weights. By modeling the \emph{metric}, which is the core tensor field of general relativity, Einstein Fields enable the derivation of physical quantities via automatic differentiation. However, unlike conventional neural fields (e.g., signed distance, occupancy, or radiance fields), Einstein Fields are \emph{Neural Tensor Fields} with the key difference that when encoding the spacetime geometry of general relativity into neural field representations, dynamics emerge naturally as a byproduct. Einstein Fields show remarkable potential, including continuum modeling of 4D spacetime, mesh-agnosticity, storage efficiency, derivative accuracy, and ease of use. We address these challenges across several canonical test beds of general relativity and release an open source JAX-based library, paving the way for more scalable and expressive approaches to numerical relativity. Code is made available at https://github.com/AndreiB137/EinFields
SIMSHIFT: A Benchmark for Adapting Neural Surrogates to Distribution Shifts
Jun 13, 2025Abstract:Neural surrogates for Partial Differential Equations (PDEs) often suffer significant performance degradation when evaluated on unseen problem configurations, such as novel material types or structural dimensions. Meanwhile, Domain Adaptation (DA) techniques have been widely used in vision and language processing to generalize from limited information about unseen configurations. In this work, we address this gap through two focused contributions. First, we introduce SIMSHIFT, a novel benchmark dataset and evaluation suite composed of four industrial simulation tasks: hot rolling, sheet metal forming, electric motor design and heatsink design. Second, we extend established domain adaptation methods to state of the art neural surrogates and systematically evaluate them. These approaches use parametric descriptions and ground truth simulations from multiple source configurations, together with only parametric descriptions from target configurations. The goal is to accurately predict target simulations without access to ground truth simulation data. Extensive experiments on SIMSHIFT highlight the challenges of out of distribution neural surrogate modeling, demonstrate the potential of DA in simulation, and reveal open problems in achieving robust neural surrogates under distribution shifts in industrially relevant scenarios. Our codebase is available at https://github.com/psetinek/simshift
LaM-SLidE: Latent Space Modeling of Spatial Dynamical Systems via Linked Entities
Feb 17, 2025Abstract:Generative models are spearheading recent progress in deep learning, showing strong promise for trajectory sampling in dynamical systems as well. However, while latent space modeling paradigms have transformed image and video generation, similar approaches are more difficult for most dynamical systems. Such systems -- from chemical molecule structures to collective human behavior -- are described by interactions of entities, making them inherently linked to connectivity patterns and the traceability of entities over time. Our approach, LaM-SLidE (Latent Space Modeling of Spatial Dynamical Systems via Linked Entities), combines the advantages of graph neural networks, i.e., the traceability of entities across time-steps, with the efficiency and scalability of recent advances in image and video generation, where pre-trained encoder and decoder are frozen to enable generative modeling in the latent space. The core idea of LaM-SLidE is to introduce identifier representations (IDs) to allow for retrieval of entity properties, e.g., entity coordinates, from latent system representations and thus enables traceability. Experimentally, across different domains, we show that LaM-SLidE performs favorably in terms of speed, accuracy, and generalizability. (Code is available at https://github.com/ml-jku/LaM-SLidE)
Generative Topology Optimization: Exploring Diverse Solutions in Structural Design
Feb 17, 2025



Abstract:Topology optimization (TO) is a family of computational methods that derive near-optimal geometries from formal problem descriptions. Despite their success, established TO methods are limited to generating single solutions, restricting the exploration of alternative designs. To address this limitation, we introduce Generative Topology Optimization (GenTO) - a data-free method that trains a neural network to generate structurally compliant shapes and explores diverse solutions through an explicit diversity constraint. The network is trained with a solver-in-the-loop, optimizing the material distribution in each iteration. The trained model produces diverse shapes that closely adhere to the design requirements. We validate GenTO on 2D and 3D TO problems. Our results demonstrate that GenTO produces more diverse solutions than any prior method while maintaining near-optimality and being an order of magnitude faster due to inherent parallelism. These findings open new avenues for engineering and design, offering enhanced flexibility and innovation in structural optimization.
5D Neural Surrogates for Nonlinear Gyrokinetic Simulations of Plasma Turbulence
Feb 11, 2025Abstract:Nuclear fusion plays a pivotal role in the quest for reliable and sustainable energy production. A major roadblock to achieving commercially viable fusion power is understanding plasma turbulence, which can significantly degrade plasma confinement. Modelling turbulence is crucial to design performing plasma scenarios for next-generation reactor-class devices and current experimental machines. The nonlinear gyrokinetic equation underpinning turbulence modelling evolves a 5D distribution function over time. Solving this equation numerically is extremely expensive, requiring up to weeks for a single run to converge, making it unfeasible for iterative optimisation and control studies. In this work, we propose a method for training neural surrogates for 5D gyrokinetic simulations. Our method extends a hierarchical vision transformer to five dimensions and is trained on the 5D distribution function for the adiabatic electron approximation. We demonstrate that our model can accurately infer downstream physical quantities such as heat flux time trace and electrostatic potentials for single-step predictions two orders of magnitude faster than numerical codes. Our work paves the way towards neural surrogates for plasma turbulence simulations to accelerate deployment of commercial energy production via nuclear fusion.
NeuralDEM -- Real-time Simulation of Industrial Particulate Flows
Nov 14, 2024Abstract:Advancements in computing power have made it possible to numerically simulate large-scale fluid-mechanical and/or particulate systems, many of which are integral to core industrial processes. Among the different numerical methods available, the discrete element method (DEM) provides one of the most accurate representations of a wide range of physical systems involving granular and discontinuous materials. Consequently, DEM has become a widely accepted approach for tackling engineering problems connected to granular flows and powder mechanics. Additionally, DEM can be integrated with grid-based computational fluid dynamics (CFD) methods, enabling the simulation of chemical processes taking place, e.g., in fluidized beds. However, DEM is computationally intensive because of the intrinsic multiscale nature of particulate systems, restricting simulation duration or number of particles. Towards this end, NeuralDEM presents an end-to-end approach to replace slow numerical DEM routines with fast, adaptable deep learning surrogates. NeuralDEM is capable of picturing long-term transport processes across different regimes using macroscopic observables without any reference to microscopic model parameters. First, NeuralDEM treats the Lagrangian discretization of DEM as an underlying continuous field, while simultaneously modeling macroscopic behavior directly as additional auxiliary fields. Second, NeuralDEM introduces multi-branch neural operators scalable to real-time modeling of industrially-sized scenarios - from slow and pseudo-steady to fast and transient. Such scenarios have previously posed insurmountable challenges for deep learning models. Notably, NeuralDEM faithfully models coupled CFD-DEM fluidized bed reactors of 160k CFD cells and 500k DEM particles for trajectories of 28s. NeuralDEM will open many new doors to advanced engineering and much faster process cycles.
Bio-xLSTM: Generative modeling, representation and in-context learning of biological and chemical sequences
Nov 06, 2024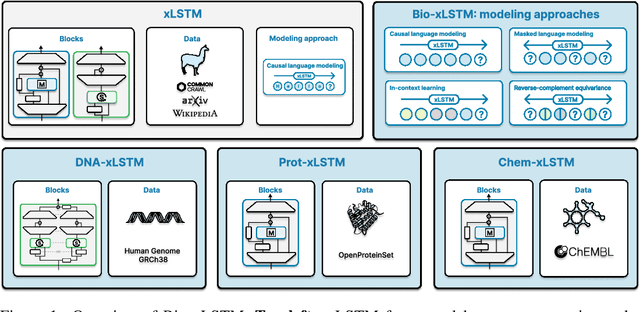
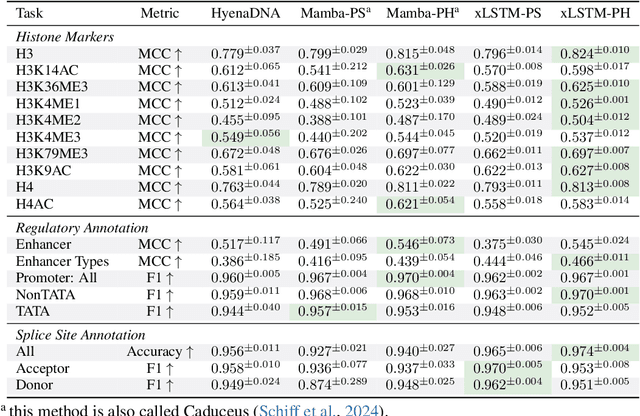
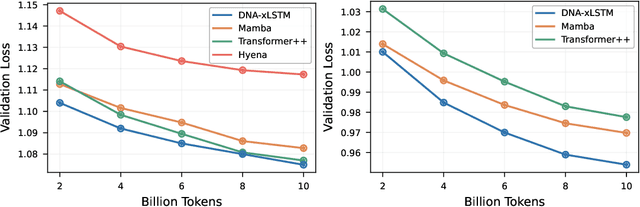
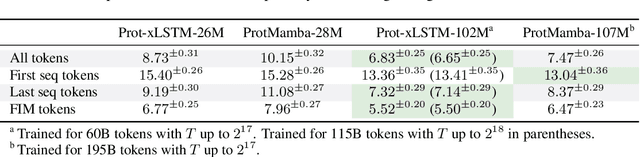
Abstract:Language models for biological and chemical sequences enable crucial applications such as drug discovery, protein engineering, and precision medicine. Currently, these language models are predominantly based on Transformer architectures. While Transformers have yielded impressive results, their quadratic runtime dependency on the sequence length complicates their use for long genomic sequences and in-context learning on proteins and chemical sequences. Recently, the recurrent xLSTM architecture has been shown to perform favorably compared to Transformers and modern state-space model (SSM) architectures in the natural language domain. Similar to SSMs, xLSTMs have a linear runtime dependency on the sequence length and allow for constant-memory decoding at inference time, which makes them prime candidates for modeling long-range dependencies in biological and chemical sequences. In this work, we tailor xLSTM towards these domains and propose a suite of architectural variants called Bio-xLSTM. Extensive experiments in three large domains, genomics, proteins, and chemistry, were performed to assess xLSTM's ability to model biological and chemical sequences. The results show that models based on Bio-xLSTM a) can serve as proficient generative models for DNA, protein, and chemical sequences, b) learn rich representations for those modalities, and c) can perform in-context learning for proteins and small molecules.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge