Matthias Schmid
Institute for Medical Biometry, Informatics and Epidemiology, University of Bonn
Characterizing gaussian mixture of motion modes for skid-steer state estimation
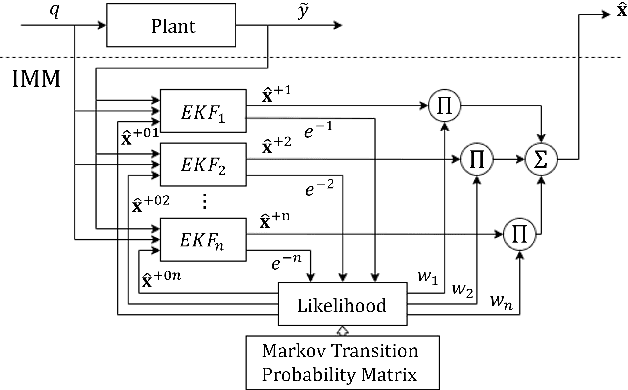
Apr 30, 2025Abstract:Skid-steered wheel mobile robots (SSWMRs) are characterized by the unique domination of the tire-terrain skidding for the robot to move. The lack of reliable friction models cascade into unreliable motion models, especially the reduced ordered variants used for state estimation and robot control. Ensemble modeling is an emerging research direction where the overall motion model is broken down into a family of local models to distribute the performance and resource requirement and provide a fast real-time prediction. To this end, a gaussian mixture model based modeling identification of model clusters is adopted and implemented within an interactive multiple model (IMM) based state estimation. The framework is adopted and implemented for angular velocity as the estimated state for a mid scaled skid-steered wheel mobile robot platform.
Online identification of skidding modes with interactive multiple model estimation
Sep 30, 2024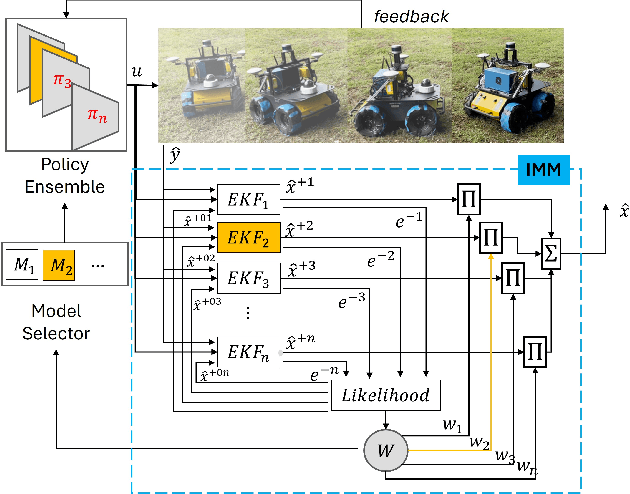
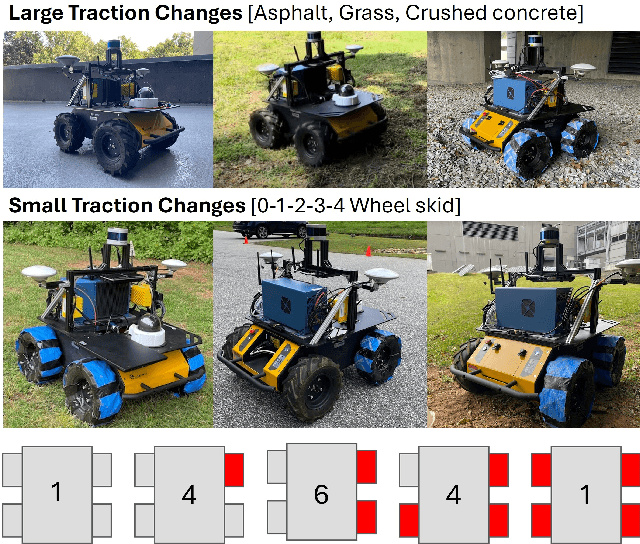
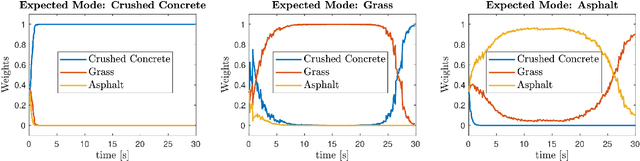
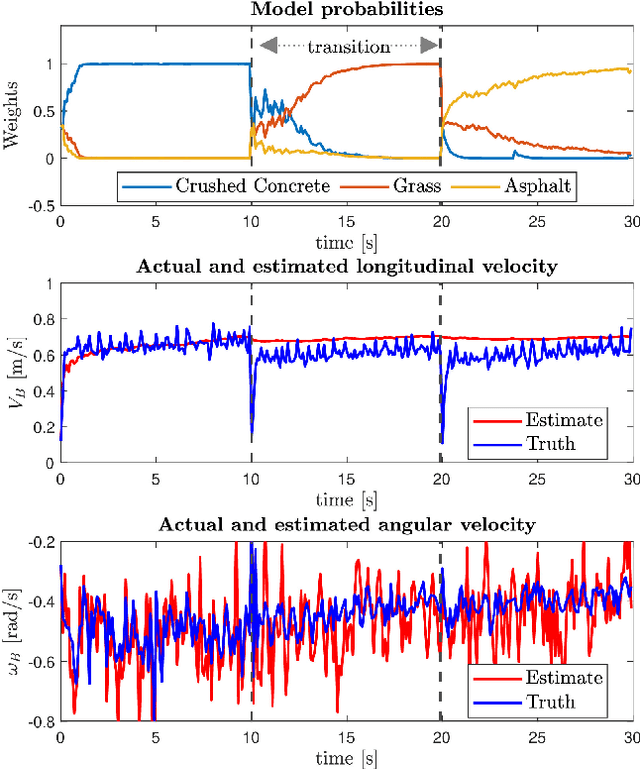
Abstract:Skid-steered wheel mobile robots (SSWMRs) operate in a variety of outdoor environments exhibiting motion behaviors dominated by the effects of complex wheel-ground interactions. Characterizing these interactions is crucial both from the immediate robot autonomy perspective (for motion prediction and control) as well as a long-term predictive maintenance and diagnostics perspective. An ideal solution entails capturing precise state measurements for decisions and controls, which is considerably difficult, especially in increasingly unstructured outdoor regimes of operations for these robots. In this milieu, a framework to identify pre-determined discrete modes of operation can considerably simplify the motion model identification process. To this end, we propose an interactive multiple model (IMM) based filtering framework to probabilistically identify predefined robot operation modes that could arise due to traversal in different terrains or loss of wheel traction.
Achieving interpretable machine learning by functional decomposition of black-box models into explainable predictor effects
Jul 26, 2024Abstract:Machine learning (ML) has seen significant growth in both popularity and importance. The high prediction accuracy of ML models is often achieved through complex black-box architectures that are difficult to interpret. This interpretability problem has been hindering the use of ML in fields like medicine, ecology and insurance, where an understanding of the inner workings of the model is paramount to ensure user acceptance and fairness. The need for interpretable ML models has boosted research in the field of interpretable machine learning (IML). Here we propose a novel approach for the functional decomposition of black-box predictions, which is considered a core concept of IML. The idea of our method is to replace the prediction function by a surrogate model consisting of simpler subfunctions. Similar to additive regression models, these functions provide insights into the direction and strength of the main feature contributions and their interactions. Our method is based on a novel concept termed stacked orthogonality, which ensures that the main effects capture as much functional behavior as possible and do not contain information explained by higher-order interactions. Unlike earlier functional IML approaches, it is neither affected by extrapolation nor by hidden feature interactions. To compute the subfunctions, we propose an algorithm based on neural additive modeling and an efficient post-hoc orthogonalization procedure.
Model-based recursive partitioning for discrete event times
Sep 14, 2022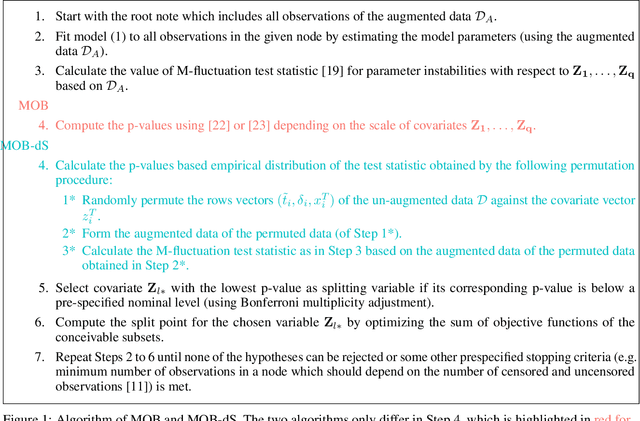

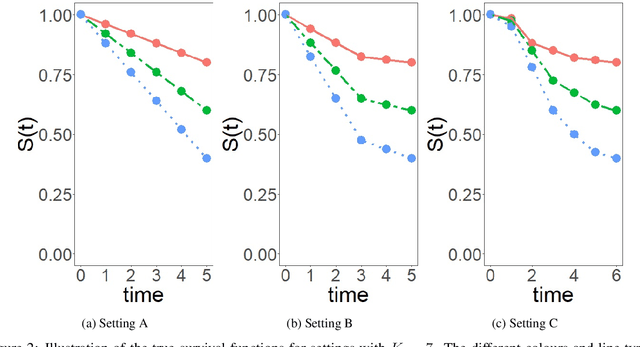
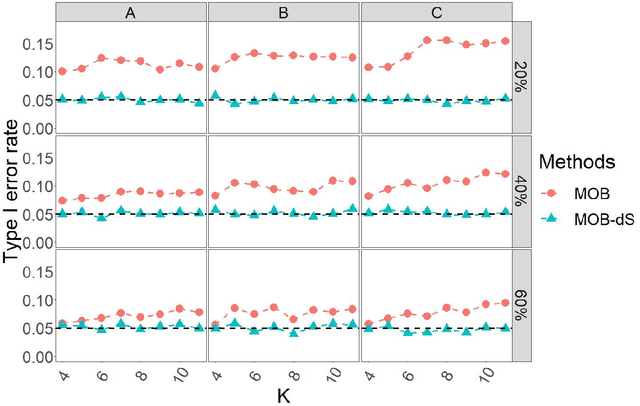
Abstract:Model-based recursive partitioning (MOB) is a semi-parametric statistical approach allowing the identification of subgroups that can be combined with a broad range of outcome measures including continuous time-to-event outcomes. When time is measured on a discrete scale, methods and models need to account for this discreetness as otherwise subgroups might be spurious and effects biased. The test underlying the splitting criterion of MOB, the M-fluctuation test, assumes independent observations. However, for fitting discrete time-to-event models the data matrix has to be modified resulting in an augmented data matrix violating the independence assumption. We propose MOB for discrete Survival data (MOB-dS) which controls the type I error rate of the test used for data splitting and therefore the rate of identifying subgroups although none is present. MOB-ds uses a permutation approach accounting for dependencies in the augmented time-to-event data to obtain the distribution under the null hypothesis of no subgroups being present. Through simulations we investigate the type I error rate of the new MOB-dS and the standard MOB for different patterns of survival curves and event rates. We find that the type I error rates of the test is well controlled for MOB-dS, but observe some considerable inflations of the error rate for MOB. To illustrate the proposed methods, MOB-dS is applied to data on unemployment duration.
A Failure Identification and Recovery Framework for a Planar Reconfigurable Cable Driven Parallel Robot
Sep 02, 2022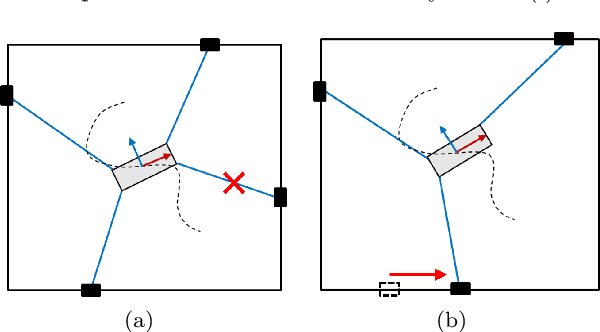
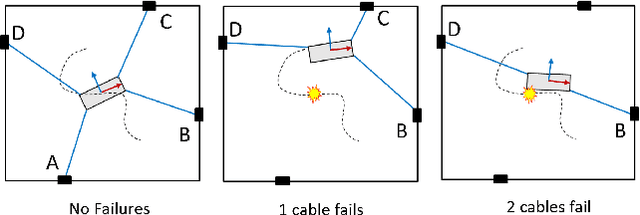
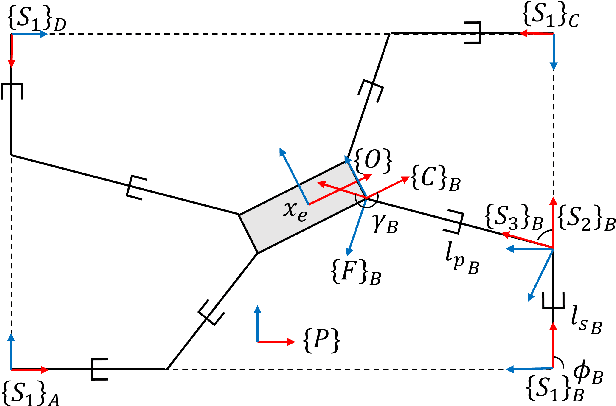
Abstract:In cable driven parallel robots (CDPRs), a single cable malfunction usually induces complete failure of the entire robot. However, the lost static workspace (due to failure) can often be recovered through reconfiguration of the cable attachment points on the frame. This capability is introduced by adding kinematic redundancies to the robot in the form of moving linear sliders that are manipulated in a real-time redundancy resolution controller. The presented work combines this controller with an online failure detection framework to develop a complete fault tolerant control scheme for automatic task recovery. This solution provides robustness by combining pose estimation of the end-effector with the failure detection through the application of an Interactive Multiple Model (IMM) algorithm relying only on end-effector information. The failure and pose estimation scheme is then tied into the redundancy resolution approach to produce a seamless automatic task (trajectory) recovery approach for cable failures.
Total Least Squares for Optimal Pose Estimation
Jun 22, 2021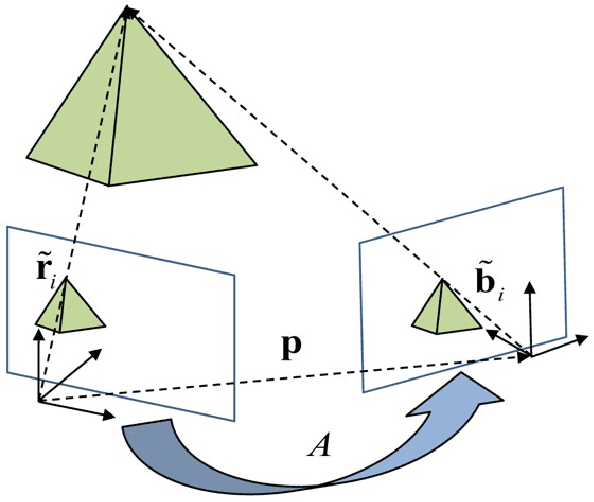
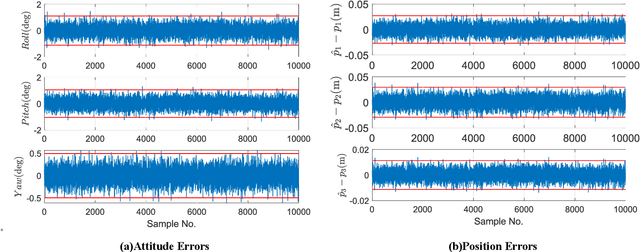
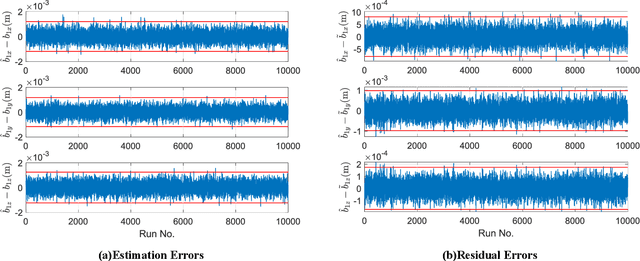
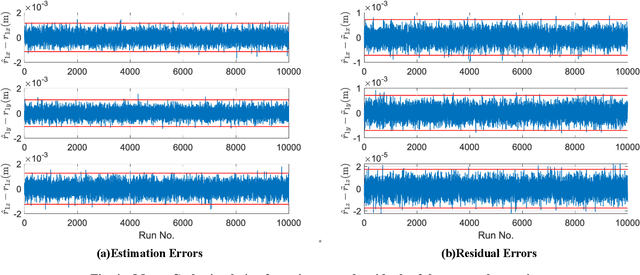
Abstract:This work provides a theoretical framework for the pose estimation problem using total least squares for vector observations from landmark features. First, the optimization framework is formulated for the pose estimation problem with observation vectors extracted from point cloud features. Then, error-covariance expressions are derived. The attitude and position solutions obtained via the derived optimization framework are proven to reach the bounds defined by the Cram\'er-Rao lower bound under the small angle approximation of attitude errors. The measurement data for the simulation of this problem is provided through a series of vector observation scans, and a fully populated observation noise-covariance matrix is assumed as the weight in the cost function to cover for the most general case of the sensor uncertainty. Here, previous derivations are expanded for the pose estimation problem to include more generic cases of correlations in the errors than previously cases involving an isotropic noise assumption. The proposed solution is simulated in a Monte-Carlo framework with 10,000 samples to validate the error-covariance analysis.
An update on statistical boosting in biomedicine
Feb 27, 2017
Abstract:Statistical boosting algorithms have triggered a lot of research during the last decade. They combine a powerful machine-learning approach with classical statistical modelling, offering various practical advantages like automated variable selection and implicit regularization of effect estimates. They are extremely flexible, as the underlying base-learners (regression functions defining the type of effect for the explanatory variables) can be combined with any kind of loss function (target function to be optimized, defining the type of regression setting). In this review article, we highlight the most recent methodological developments on statistical boosting regarding variable selection, functional regression and advanced time-to-event modelling. Additionally, we provide a short overview on relevant applications of statistical boosting in biomedicine.
Boosting Joint Models for Longitudinal and Time-to-Event Data
Dec 22, 2016



Abstract:Joint Models for longitudinal and time-to-event data have gained a lot of attention in the last few years as they are a helpful technique to approach common a data structure in clinical studies where longitudinal outcomes are recorded alongside event times. Those two processes are often linked and the two outcomes should thus be modeled jointly in order to prevent the potential bias introduced by independent modelling. Commonly, joint models are estimated in likelihood based expectation maximization or Bayesian approaches using frameworks where variable selection is problematic and which do not immediately work for high-dimensional data. In this paper, we propose a boosting algorithm tackling these challenges by being able to simultaneously estimate predictors for joint models and automatically select the most influential variables even in high-dimensional data situations. We analyse the performance of the new algorithm in a simulation study and apply it to the Danish cystic fibrosis registry which collects longitudinal lung function data on patients with cystic fibrosis together with data regarding the onset of pulmonary infections. This is the first approach to combine state-of-the art algorithms from the field of machine-learning with the model class of joint models, providing a fully data-driven mechanism to select variables and predictor effects in a unified framework of boosting joint models.
Stability selection for component-wise gradient boosting in multiple dimensions
Nov 30, 2016



Abstract:We present a new algorithm for boosting generalized additive models for location, scale and shape (GAMLSS) that allows to incorporate stability selection, an increasingly popular way to obtain stable sets of covariates while controlling the per-family error rate (PFER). The model is fitted repeatedly to subsampled data and variables with high selection frequencies are extracted. To apply stability selection to boosted GAMLSS, we develop a new "noncyclical" fitting algorithm that incorporates an additional selection step of the best-fitting distribution parameter in each iteration. This new algorithms has the additional advantage that optimizing the tuning parameters of boosting is reduced from a multi-dimensional to a one-dimensional problem with vastly decreased complexity. The performance of the novel algorithm is evaluated in an extensive simulation study. We apply this new algorithm to a study to estimate abundance of common eider in Massachusetts, USA, featuring excess zeros, overdispersion, non-linearity and spatio-temporal structures. Eider abundance is estimated via boosted GAMLSS, allowing both mean and overdispersion to be regressed on covariates. Stability selection is used to obtain a sparse set of stable predictors.
On the use of Harrell's C for clinical risk prediction via random survival forests
Jul 18, 2016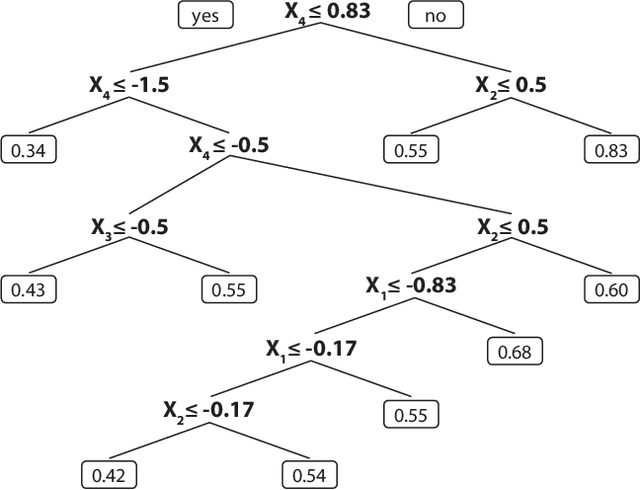
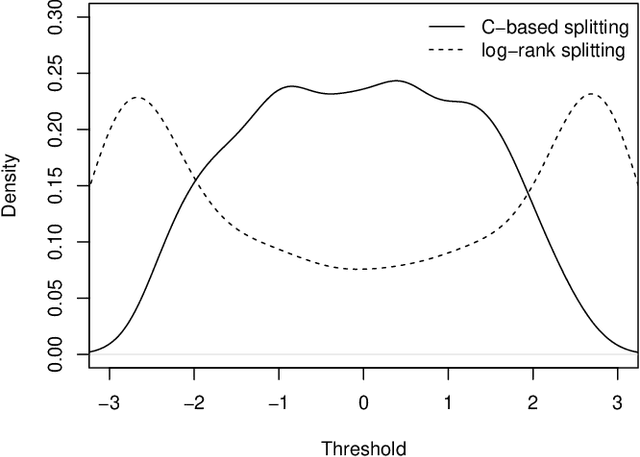
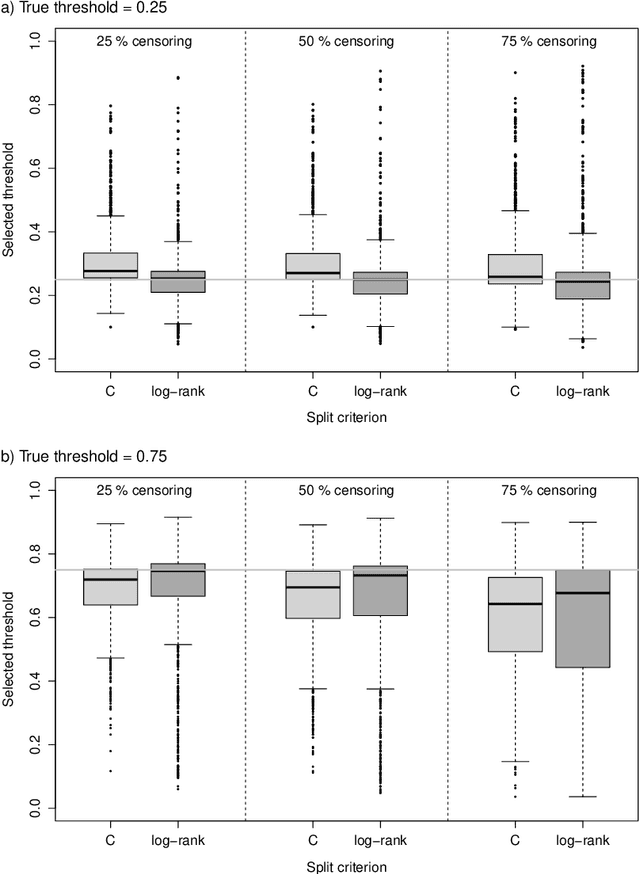
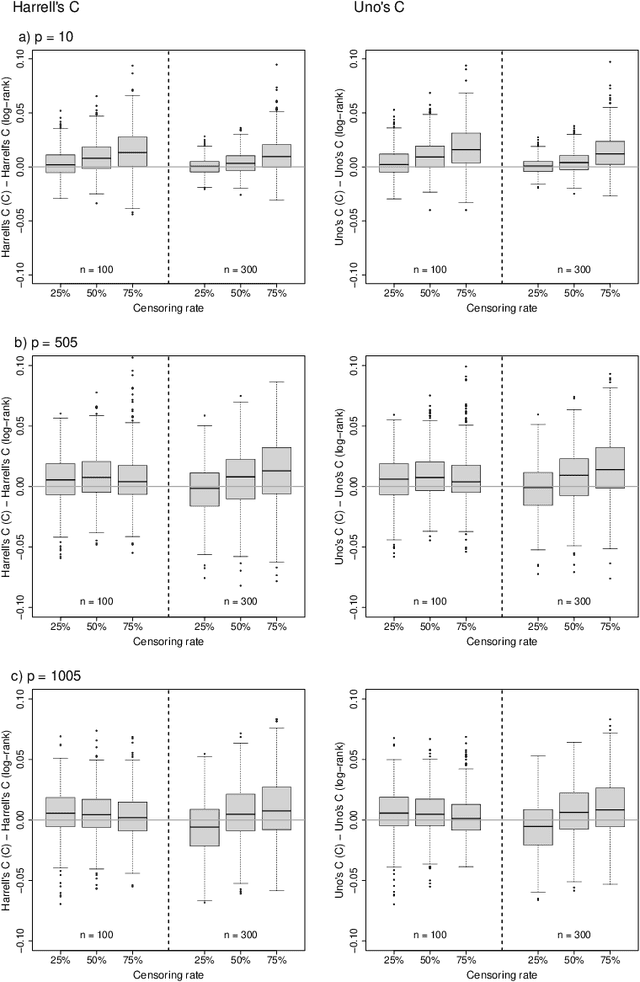
Abstract:Random survival forests (RSF) are a powerful method for risk prediction of right-censored outcomes in biomedical research. RSF use the log-rank split criterion to form an ensemble of survival trees. The most common approach to evaluate the prediction accuracy of a RSF model is Harrell's concordance index for survival data ('C index'). Conceptually, this strategy implies that the split criterion in RSF is different from the evaluation criterion of interest. This discrepancy can be overcome by using Harrell's C for both node splitting and evaluation. We compare the difference between the two split criteria analytically and in simulation studies with respect to the preference of more unbalanced splits, termed end-cut preference (ECP). Specifically, we show that the log-rank statistic has a stronger ECP compared to the C index. In simulation studies and with the help of two medical data sets we demonstrate that the accuracy of RSF predictions, as measured by Harrell's C, can be improved if the log-rank statistic is replaced by the C index for node splitting. This is especially true in situations where the censoring rate or the fraction of informative continuous predictor variables is high. Conversely, log-rank splitting is preferable in noisy scenarios. Both C-based and log-rank splitting are implemented in the R~package ranger. We recommend Harrell's C as split criterion for use in smaller scale clinical studies and the log-rank split criterion for use in large-scale 'omics' studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge