Marvin Wright
Autoencoding Random Forests
May 27, 2025
Abstract:We propose a principled method for autoencoding with random forests. Our strategy builds on foundational results from nonparametric statistics and spectral graph theory to learn a low-dimensional embedding of the model that optimally represents relationships in the data. We provide exact and approximate solutions to the decoding problem via constrained optimization, split relabeling, and nearest neighbors regression. These methods effectively invert the compression pipeline, establishing a map from the embedding space back to the input space using splits learned by the ensemble's constituent trees. The resulting decoders are universally consistent under common regularity assumptions. The procedure works with supervised or unsupervised models, providing a window into conditional or joint distributions. We demonstrate various applications of this autoencoder, including powerful new tools for visualization, compression, clustering, and denoising. Experiments illustrate the ease and utility of our method in a wide range of settings, including tabular, image, and genomic data.
CountARFactuals -- Generating plausible model-agnostic counterfactual explanations with adversarial random forests
Apr 04, 2024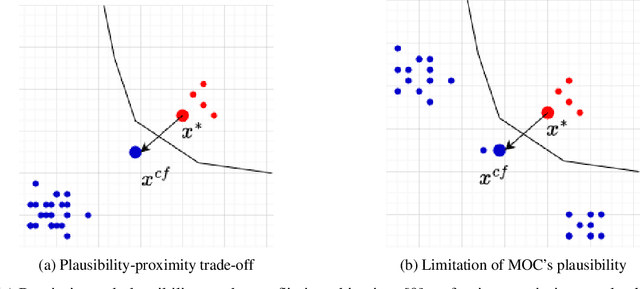
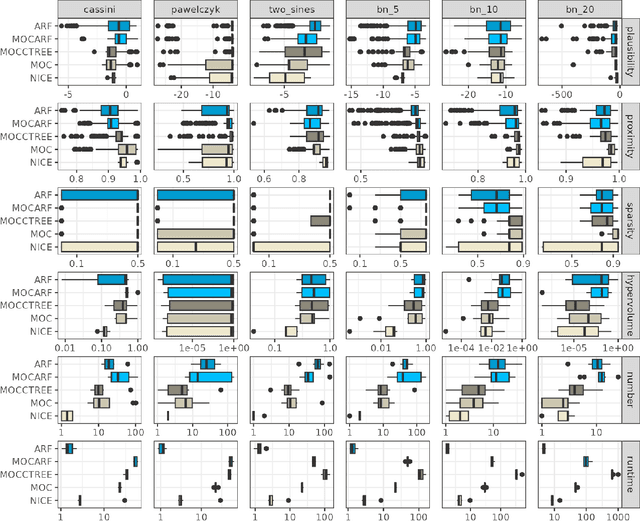

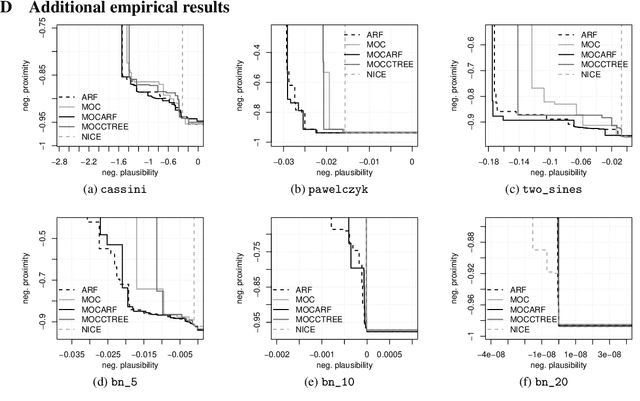
Abstract:Counterfactual explanations elucidate algorithmic decisions by pointing to scenarios that would have led to an alternative, desired outcome. Giving insight into the model's behavior, they hint users towards possible actions and give grounds for contesting decisions. As a crucial factor in achieving these goals, counterfactuals must be plausible, i.e., describing realistic alternative scenarios within the data manifold. This paper leverages a recently developed generative modeling technique -- adversarial random forests (ARFs) -- to efficiently generate plausible counterfactuals in a model-agnostic way. ARFs can serve as a plausibility measure or directly generate counterfactual explanations. Our ARF-based approach surpasses the limitations of existing methods that aim to generate plausible counterfactual explanations: It is easy to train and computationally highly efficient, handles continuous and categorical data naturally, and allows integrating additional desiderata such as sparsity in a straightforward manner.
Hyperparameters and Tuning Strategies for Random Forest
Apr 10, 2018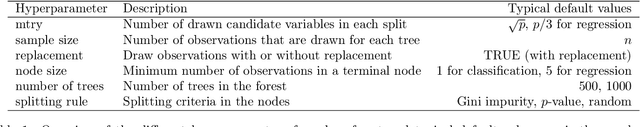
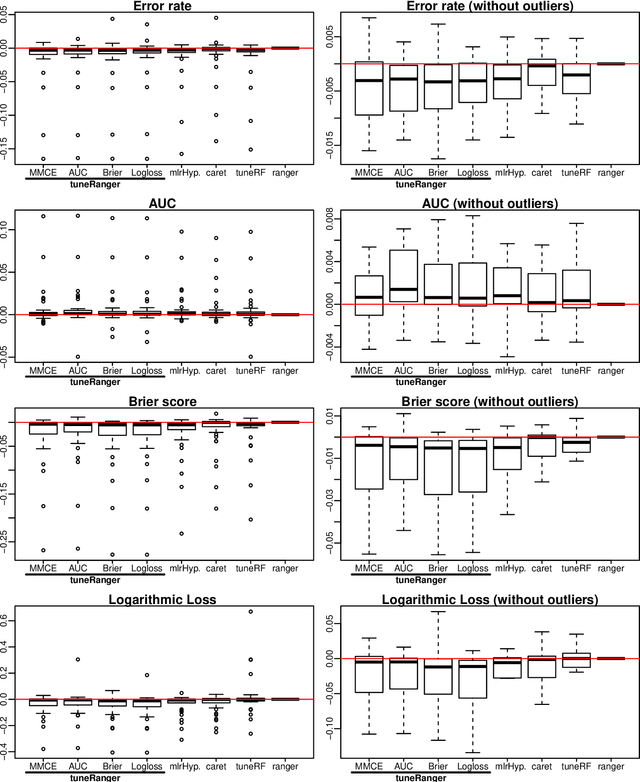
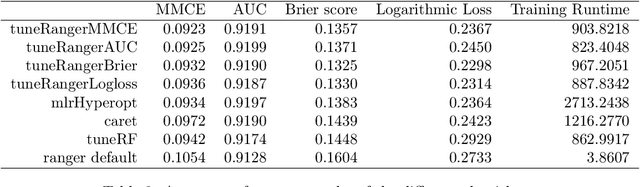
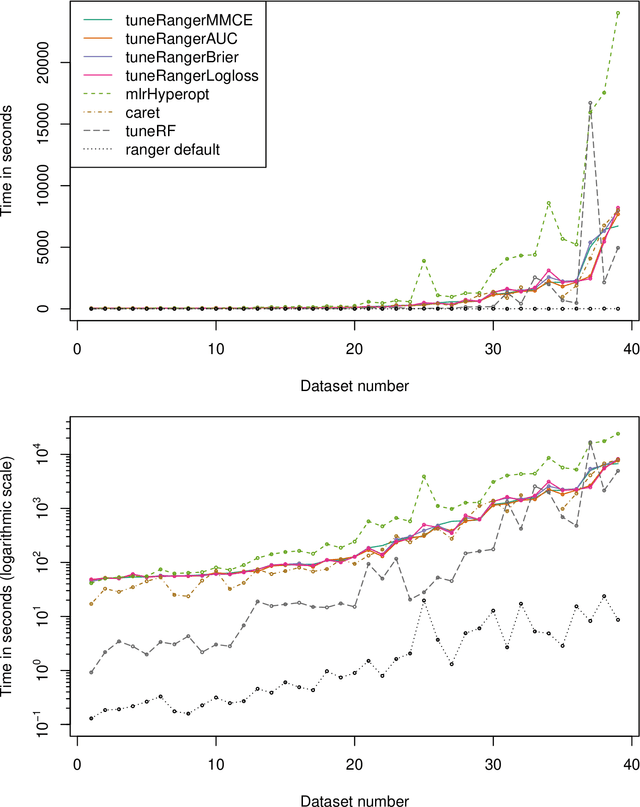
Abstract:The random forest algorithm (RF) has several hyperparameters that have to be set by the user, e.g., the number of observations drawn randomly for each tree and whether they are drawn with or without replacement, the number of variables drawn randomly for each split, the splitting rule, the minimum number of samples that a node must contain and the number of trees. In this paper, we first provide a literature review on the parameters' influence on the prediction performance and on variable importance measures, also considering interactions between hyperparameters. It is well known that in most cases RF works reasonably well with the default values of the hyperparameters specified in software packages. Nevertheless, tuning the hyperparameters can improve the performance of RF. In the second part of this paper, after a brief overview of tuning strategies we demonstrate the application of one of the most established tuning strategies, model-based optimization (MBO). To make it easier to use, we provide the tuneRanger R package that tunes RF with MBO automatically. In a benchmark study on several datasets, we compare the prediction performance and runtime of tuneRanger with other tuning implementations in R and RF with default hyperparameters.
On the use of Harrell's C for clinical risk prediction via random survival forests
Jul 18, 2016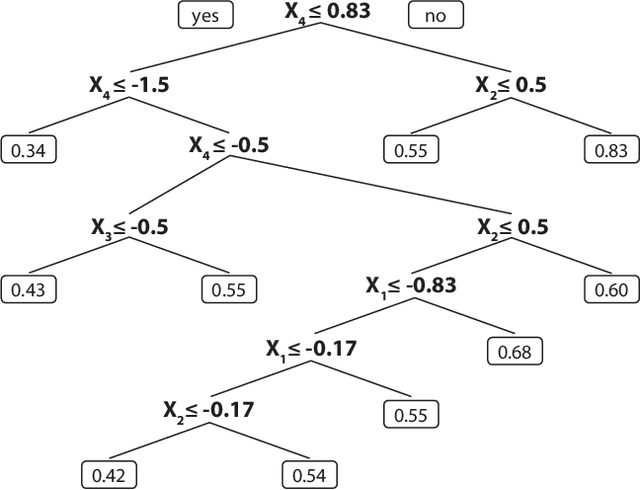
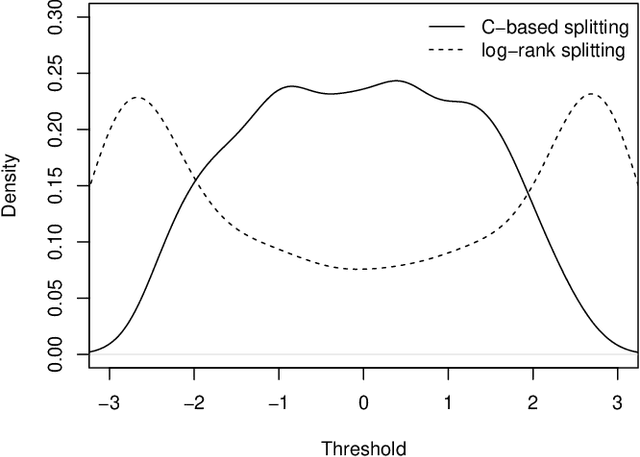
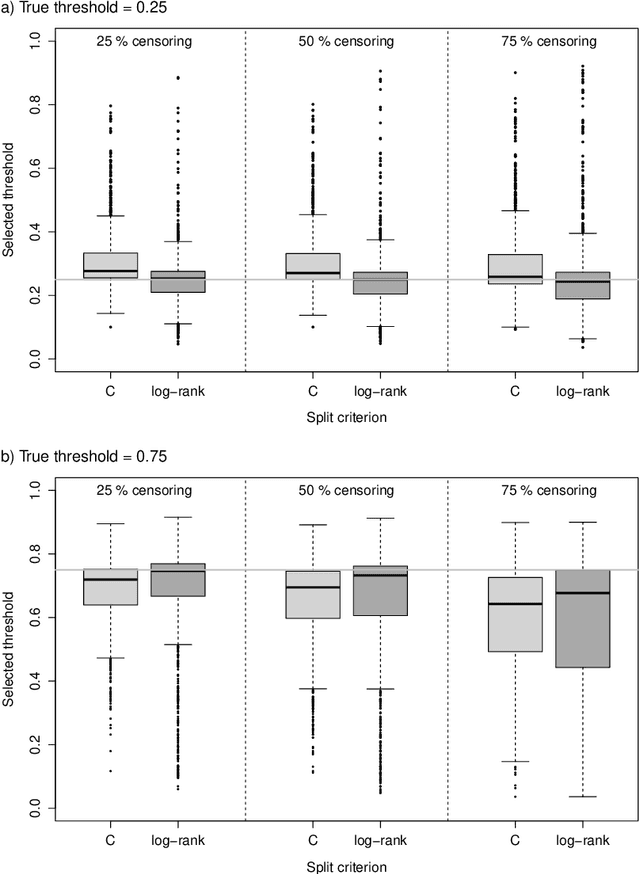
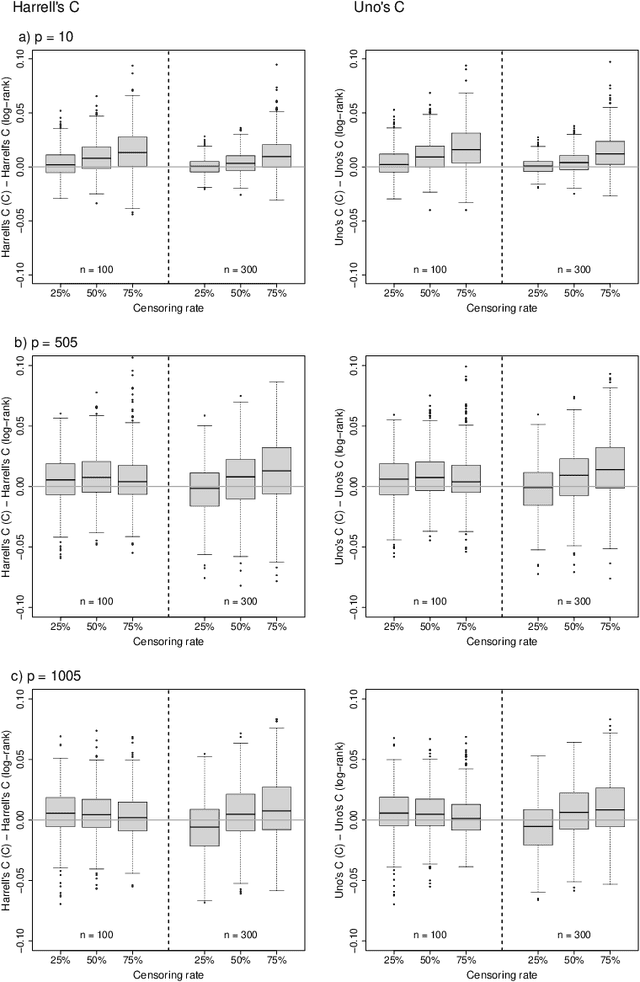
Abstract:Random survival forests (RSF) are a powerful method for risk prediction of right-censored outcomes in biomedical research. RSF use the log-rank split criterion to form an ensemble of survival trees. The most common approach to evaluate the prediction accuracy of a RSF model is Harrell's concordance index for survival data ('C index'). Conceptually, this strategy implies that the split criterion in RSF is different from the evaluation criterion of interest. This discrepancy can be overcome by using Harrell's C for both node splitting and evaluation. We compare the difference between the two split criteria analytically and in simulation studies with respect to the preference of more unbalanced splits, termed end-cut preference (ECP). Specifically, we show that the log-rank statistic has a stronger ECP compared to the C index. In simulation studies and with the help of two medical data sets we demonstrate that the accuracy of RSF predictions, as measured by Harrell's C, can be improved if the log-rank statistic is replaced by the C index for node splitting. This is especially true in situations where the censoring rate or the fraction of informative continuous predictor variables is high. Conversely, log-rank splitting is preferable in noisy scenarios. Both C-based and log-rank splitting are implemented in the R~package ranger. We recommend Harrell's C as split criterion for use in smaller scale clinical studies and the log-rank split criterion for use in large-scale 'omics' studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge