Philipp Probst
Large-scale benchmark study of survival prediction methods using multi-omics data
Mar 07, 2020



Abstract:Multi-omics data, that is, datasets containing different types of high-dimensional molecular variables (often in addition to classical clinical variables), are increasingly generated for the investigation of various diseases. Nevertheless, questions remain regarding the usefulness of multi-omics data for the prediction of disease outcomes such as survival time. It is also unclear which methods are most appropriate to derive such prediction models. We aim to give some answers to these questions by means of a large-scale benchmark study using real data. Different prediction methods from machine learning and statistics were applied on 18 multi-omics cancer datasets from the database "The Cancer Genome Atlas", containing from 35 to 1,000 observations and from 60,000 to 100,000 variables. The considered outcome was the (censored) survival time. Twelve methods based on boosting, penalized regression and random forest were compared, comprising both methods that do and that do not take the group structure of the omics variables into account. The Kaplan-Meier estimate and a Cox model using only clinical variables were used as reference methods. The methods were compared using several repetitions of 5-fold cross-validation. Uno's C-index and the integrated Brier-score served as performance metrics. The results show that, although multi-omics data can improve the prediction performance, this is not generally the case. Only the method block forest slightly outperformed the Cox model on average over all datasets. Taking into account the multi-omics structure improves the predictive performance and protects variables in low-dimensional groups - especially clinical variables - from not being included in the model. All analyses are reproducible using freely available R code.
Learning Multiple Defaults for Machine Learning Algorithms
Nov 23, 2018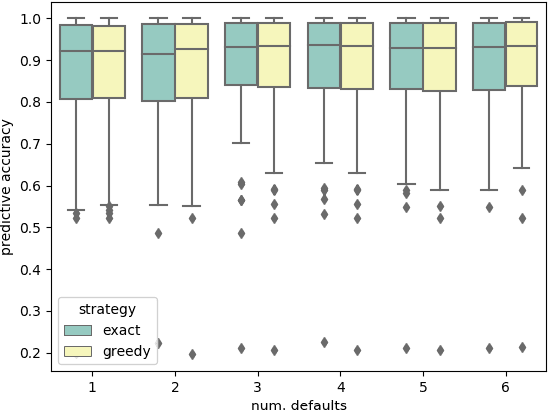
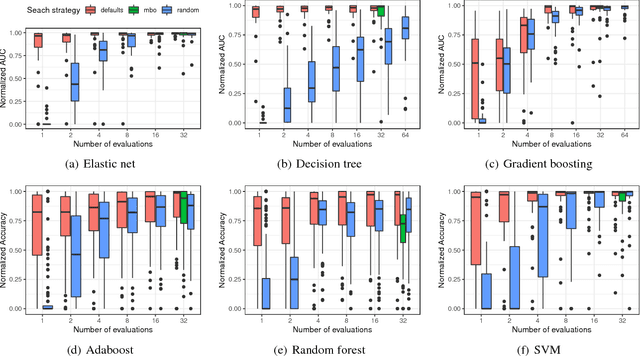
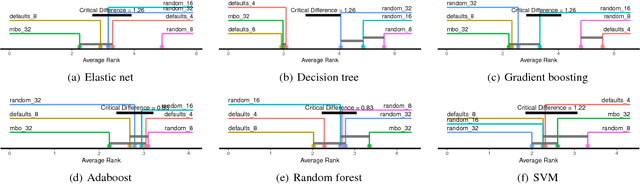
Abstract:The performance of modern machine learning methods highly depends on their hyperparameter configurations. One simple way of selecting a configuration is to use default settings, often proposed along with the publication and implementation of a new algorithm. Those default values are usually chosen in an ad-hoc manner to work good enough on a wide variety of datasets. To address this problem, different automatic hyperparameter configuration algorithms have been proposed, which select an optimal configuration per dataset. This principled approach usually improves performance, but adds additional algorithmic complexity and computational costs to the training procedure. As an alternative to this, we propose learning a set of complementary default values from a large database of prior empirical results. Selecting an appropriate configuration on a new dataset then requires only a simple, efficient and embarrassingly parallel search over this set. We demonstrate the effectiveness and efficiency of the approach we propose in comparison to random search and Bayesian Optimization.
Tunability: Importance of Hyperparameters of Machine Learning Algorithms
Oct 22, 2018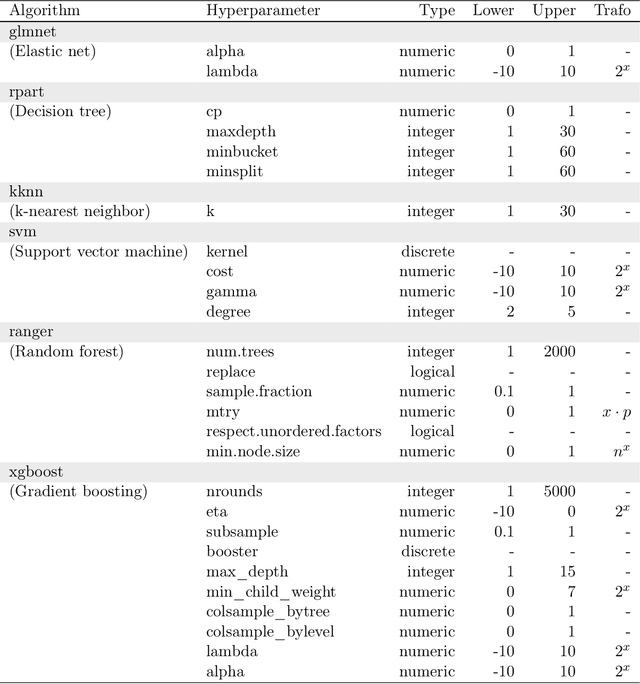
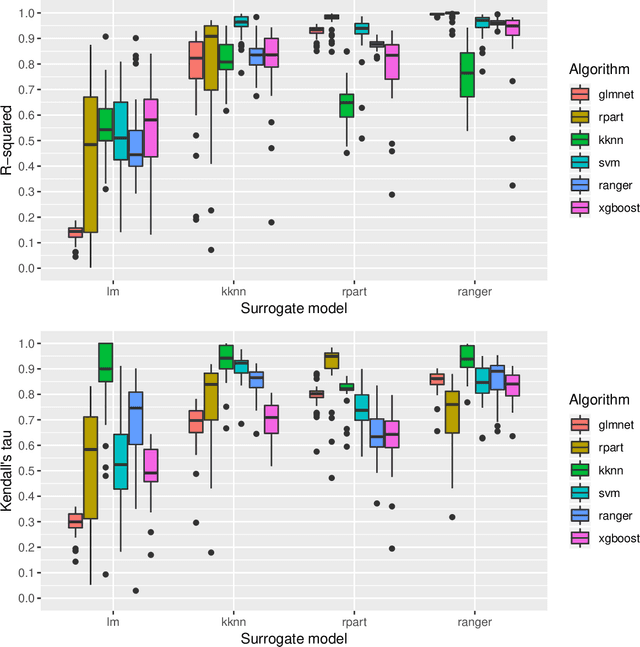
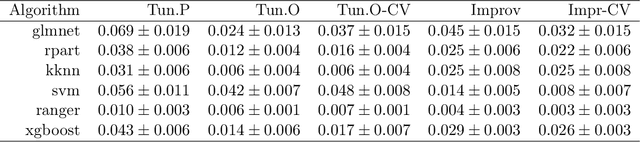
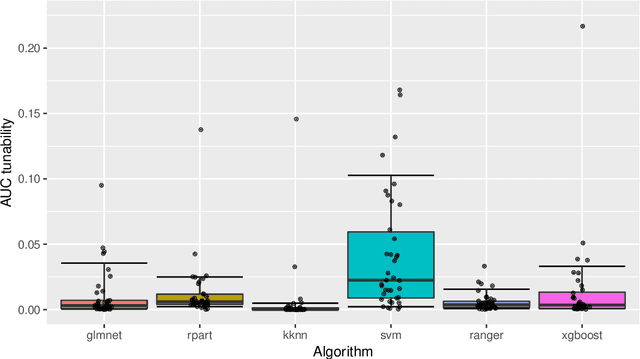
Abstract:Modern supervised machine learning algorithms involve hyperparameters that have to be set before running them. Options for setting hyperparameters are default values from the software package, manual configuration by the user or configuring them for optimal predictive performance by a tuning procedure. The goal of this paper is two-fold. Firstly, we formalize the problem of tuning from a statistical point of view, define data-based defaults and suggest general measures quantifying the tunability of hyperparameters of algorithms. Secondly, we conduct a large-scale benchmarking study based on 38 datasets from the OpenML platform and six common machine learning algorithms. We apply our measures to assess the tunability of their parameters. Our results yield default values for hyperparameters and enable users to decide whether it is worth conducting a possibly time consuming tuning strategy, to focus on the most important hyperparameters and to chose adequate hyperparameter spaces for tuning.
Automatic Exploration of Machine Learning Experiments on OpenML
Oct 19, 2018


Abstract:Understanding the influence of hyperparameters on the performance of a machine learning algorithm is an important scientific topic in itself and can help to improve automatic hyperparameter tuning procedures. Unfortunately, experimental meta data for this purpose is still rare. This paper presents a large, free and open dataset addressing this problem, containing results on 38 OpenML data sets, six different machine learning algorithms and many different hyperparameter configurations. Results where generated by an automated random sampling strategy, termed the OpenML Random Bot. Each algorithm was cross-validated up to 20.000 times per dataset with different hyperparameters settings, resulting in a meta dataset of around 2.5 million experiments overall.
Hyperparameters and Tuning Strategies for Random Forest
Apr 10, 2018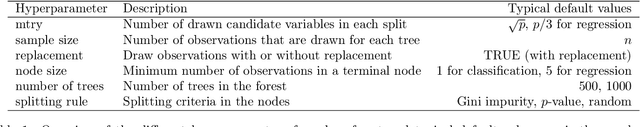
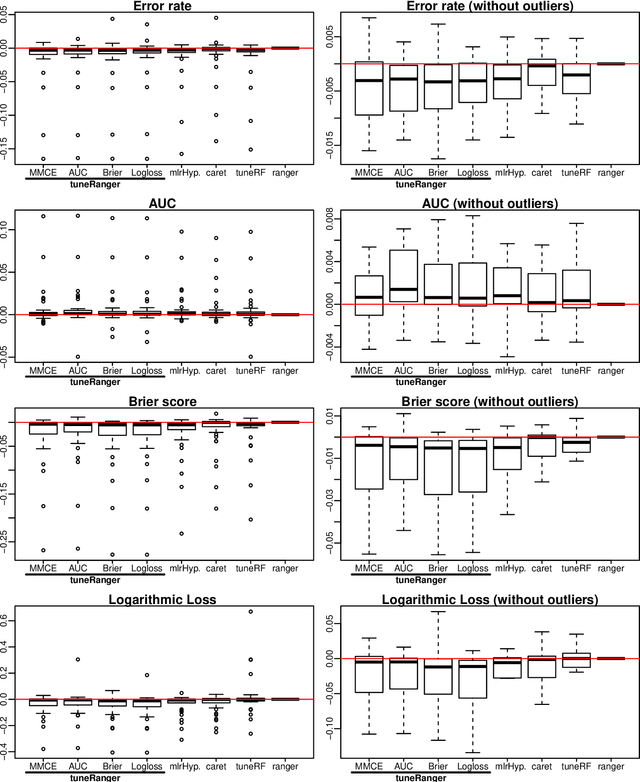
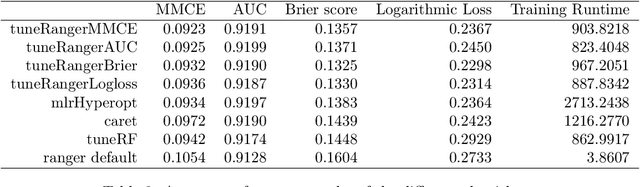
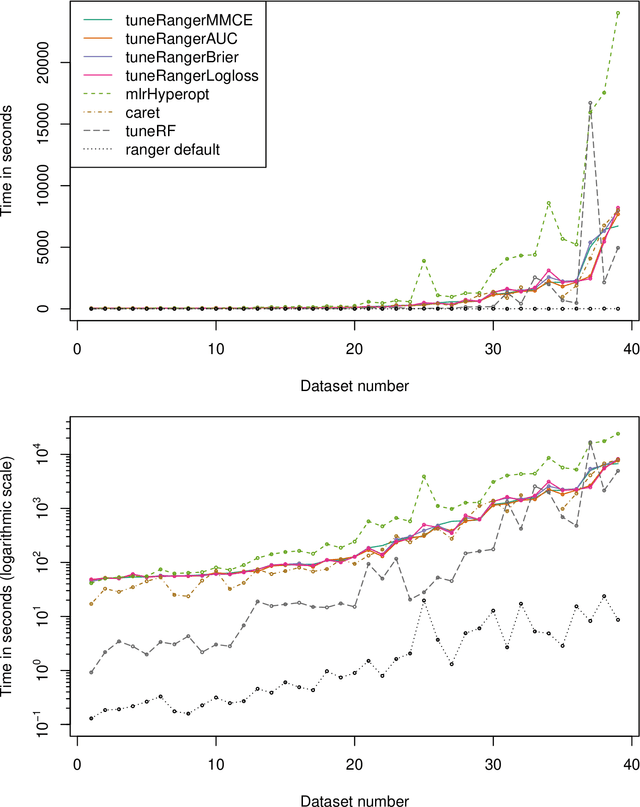
Abstract:The random forest algorithm (RF) has several hyperparameters that have to be set by the user, e.g., the number of observations drawn randomly for each tree and whether they are drawn with or without replacement, the number of variables drawn randomly for each split, the splitting rule, the minimum number of samples that a node must contain and the number of trees. In this paper, we first provide a literature review on the parameters' influence on the prediction performance and on variable importance measures, also considering interactions between hyperparameters. It is well known that in most cases RF works reasonably well with the default values of the hyperparameters specified in software packages. Nevertheless, tuning the hyperparameters can improve the performance of RF. In the second part of this paper, after a brief overview of tuning strategies we demonstrate the application of one of the most established tuning strategies, model-based optimization (MBO). To make it easier to use, we provide the tuneRanger R package that tunes RF with MBO automatically. In a benchmark study on several datasets, we compare the prediction performance and runtime of tuneRanger with other tuning implementations in R and RF with default hyperparameters.
To tune or not to tune the number of trees in random forest?
May 16, 2017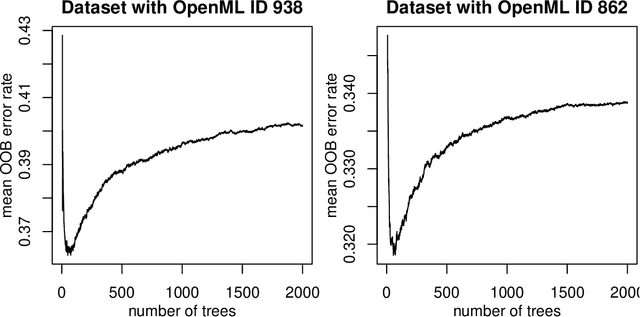
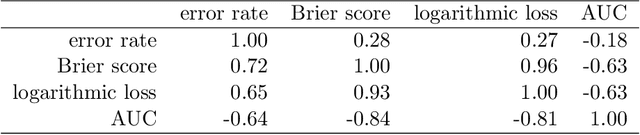
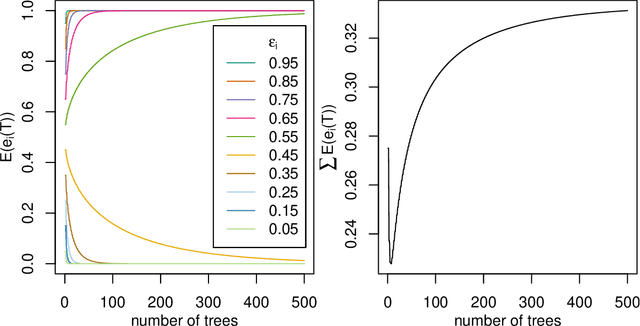
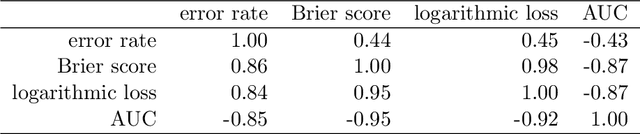
Abstract:The number of trees T in the random forest (RF) algorithm for supervised learning has to be set by the user. It is controversial whether T should simply be set to the largest computationally manageable value or whether a smaller T may in some cases be better. While the principle underlying bagging is that "more trees are better", in practice the classification error rate sometimes reaches a minimum before increasing again for increasing number of trees. The goal of this paper is four-fold: (i) providing theoretical results showing that the expected error rate may be a non-monotonous function of the number of trees and explaining under which circumstances this happens; (ii) providing theoretical results showing that such non-monotonous patterns cannot be observed for other performance measures such as the Brier score and the logarithmic loss (for classification) and the mean squared error (for regression); (iii) illustrating the extent of the problem through an application to a large number (n = 306) of datasets from the public database OpenML; (iv) finally arguing in favor of setting it to a computationally feasible large number, depending on convergence properties of the desired performance measure.
Multilabel Classification with R Package mlr
Apr 03, 2017

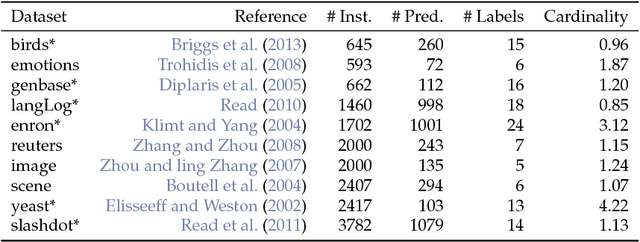

Abstract:We implemented several multilabel classification algorithms in the machine learning package mlr. The implemented methods are binary relevance, classifier chains, nested stacking, dependent binary relevance and stacking, which can be used with any base learner that is accessible in mlr. Moreover, there is access to the multilabel classification versions of randomForestSRC and rFerns. All these methods can be easily compared by different implemented multilabel performance measures and resampling methods in the standardized mlr framework. In a benchmark experiment with several multilabel datasets, the performance of the different methods is evaluated.
* 18 pages, 2 figures, to be published in R Journal; reference corrected
mlr Tutorial
Sep 18, 2016Abstract:This document provides and in-depth introduction to the mlr framework for machine learning experiments in R.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge