Moritz Herrmann
Position Paper: Rethinking Empirical Research in Machine Learning: Addressing Epistemic and Methodological Challenges of Experimentation
May 03, 2024Abstract:We warn against a common but incomplete understanding of empirical research in machine learning (ML) that leads to non-replicable results, makes findings unreliable, and threatens to undermine progress in the field. To overcome this alarming situation, we call for more awareness of the plurality of ways of gaining knowledge experimentally but also of some epistemic limitations. In particular, we argue most current empirical ML research is fashioned as confirmatory research while it should rather be considered exploratory.
DCSI -- An improved measure of cluster separability based on separation and connectedness
Oct 19, 2023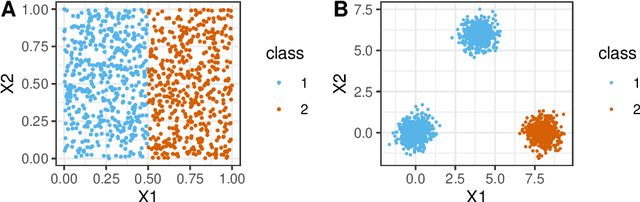
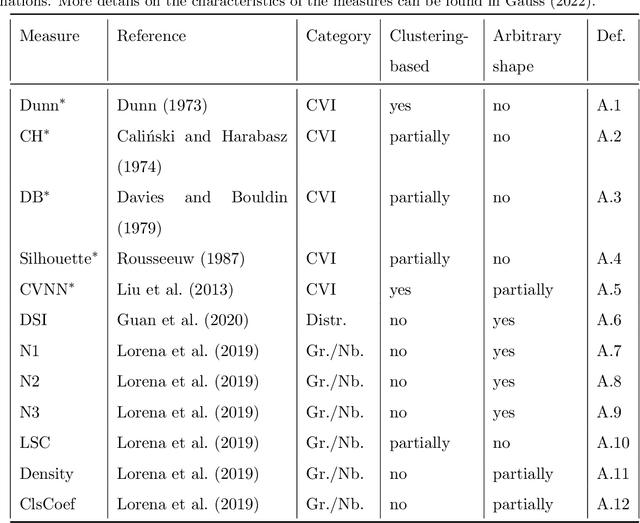
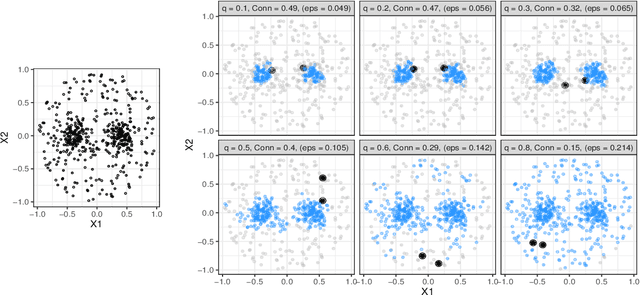
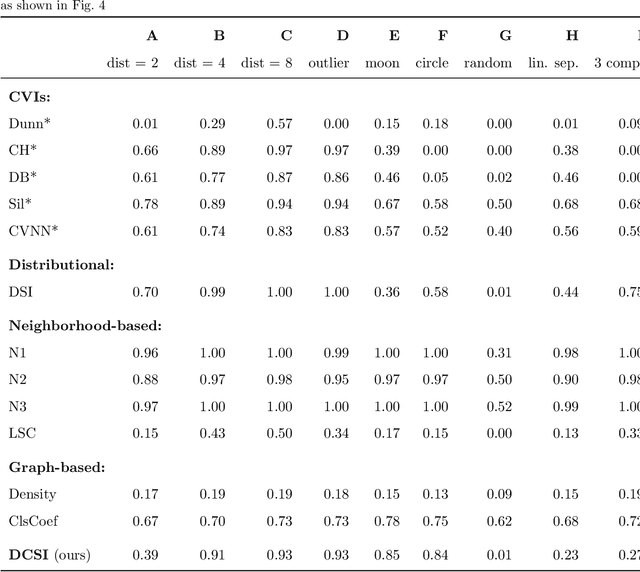
Abstract:Whether class labels in a given data set correspond to meaningful clusters is crucial for the evaluation of clustering algorithms using real-world data sets. This property can be quantified by separability measures. A review of the existing literature shows that neither classification-based complexity measures nor cluster validity indices (CVIs) adequately incorporate the central aspects of separability for density-based clustering: between-class separation and within-class connectedness. A newly developed measure (density cluster separability index, DCSI) aims to quantify these two characteristics and can also be used as a CVI. Extensive experiments on synthetic data indicate that DCSI correlates strongly with the performance of DBSCAN measured via the adjusted rand index (ARI) but lacks robustness when it comes to multi-class data sets with overlapping classes that are ill-suited for density-based hard clustering. Detailed evaluation on frequently used real-world data sets shows that DCSI can correctly identify touching or overlapping classes that do not form meaningful clusters.
Enhancing cluster analysis via topological manifold learning
Jul 01, 2022
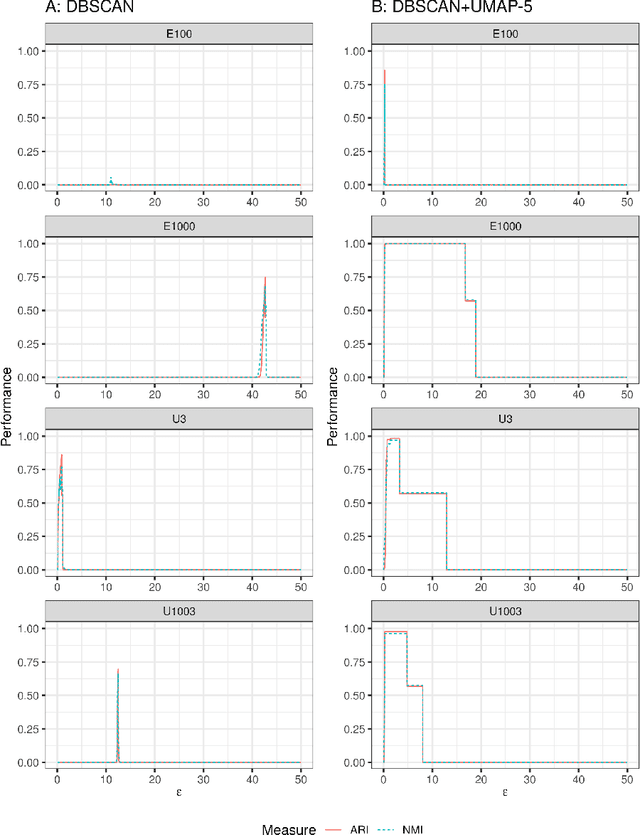
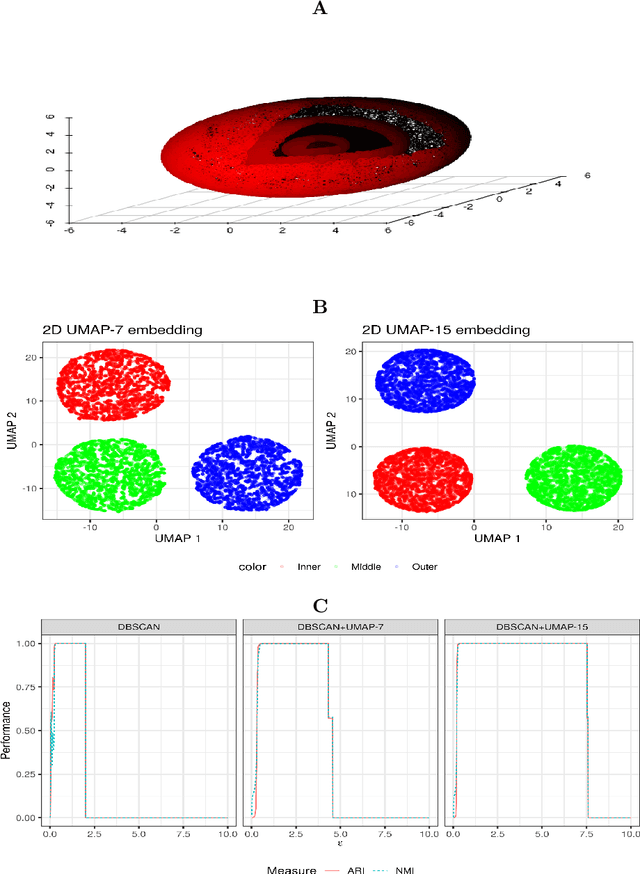

Abstract:We discuss topological aspects of cluster analysis and show that inferring the topological structure of a dataset before clustering it can considerably enhance cluster detection: theoretical arguments and empirical evidence show that clustering embedding vectors, representing the structure of a data manifold instead of the observed feature vectors themselves, is highly beneficial. To demonstrate, we combine manifold learning method UMAP for inferring the topological structure with density-based clustering method DBSCAN. Synthetic and real data results show that this both simplifies and improves clustering in a diverse set of low- and high-dimensional problems including clusters of varying density and/or entangled shapes. Our approach simplifies clustering because topological pre-processing consistently reduces parameter sensitivity of DBSCAN. Clustering the resulting embeddings with DBSCAN can then even outperform complex methods such as SPECTACL and ClusterGAN. Finally, our investigation suggests that the crucial issue in clustering does not appear to be the nominal dimension of the data or how many irrelevant features it contains, but rather how \textit{separable} the clusters are in the ambient observation space they are embedded in, which is usually the (high-dimensional) Euclidean space defined by the features of the data. Our approach is successful because we perform the cluster analysis after projecting the data into a more suitable space that is optimized for separability, in some sense.
A geometric framework for outlier detection in high-dimensional data
Jul 01, 2022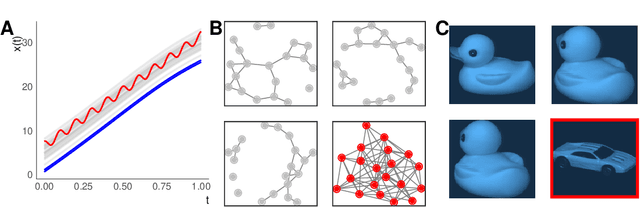
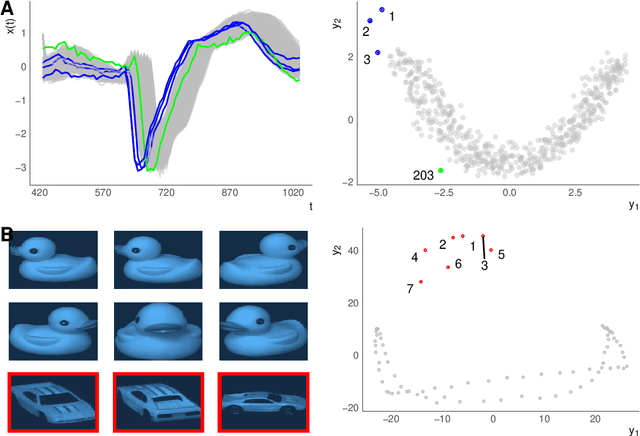
Abstract:Outlier or anomaly detection is an important task in data analysis. We discuss the problem from a geometrical perspective and provide a framework that exploits the metric structure of a data set. Our approach rests on the manifold assumption, i.e., that the observed, nominally high-dimensional data lie on a much lower dimensional manifold and that this intrinsic structure can be inferred with manifold learning methods. We show that exploiting this structure significantly improves the detection of outlying observations in high-dimensional data. We also suggest a novel, mathematically precise, and widely applicable distinction between distributional and structural outliers based on the geometry and topology of the data manifold that clarifies conceptual ambiguities prevalent throughout the literature. Our experiments focus on functional data as one class of structured high-dimensional data, but the framework we propose is completely general and we include image and graph data applications. Our results show that the outlier structure of high-dimensional and non-tabular data can be detected and visualized using manifold learning methods and quantified using standard outlier scoring methods applied to the manifold embedding vectors.
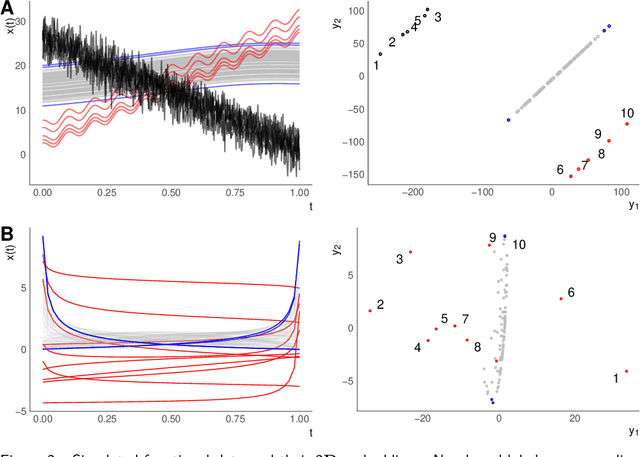
A geometric perspective on functional outlier detection
Sep 14, 2021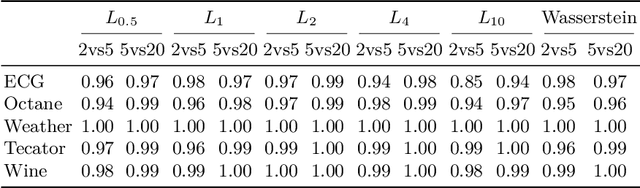
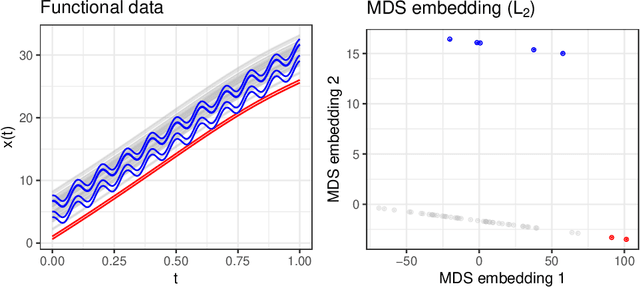
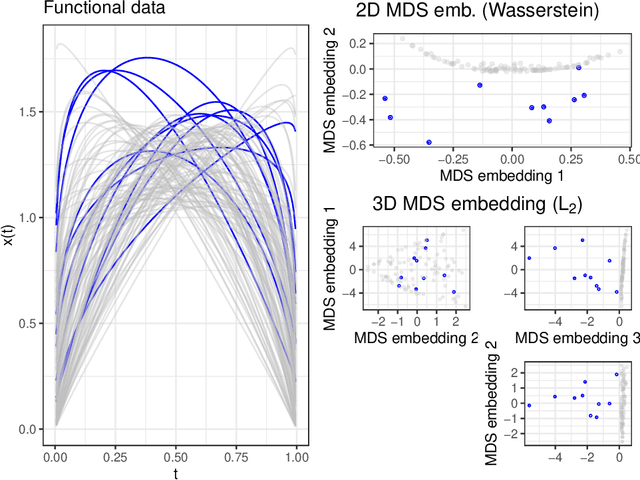
Abstract:We consider functional outlier detection from a geometric perspective, specifically: for functional data sets drawn from a functional manifold which is defined by the data's modes of variation in amplitude and phase. Based on this manifold, we develop a conceptualization of functional outlier detection that is more widely applicable and realistic than previously proposed. Our theoretical and experimental analyses demonstrate several important advantages of this perspective: It considerably improves theoretical understanding and allows to describe and analyse complex functional outlier scenarios consistently and in full generality, by differentiating between structurally anomalous outlier data that are off-manifold and distributionally outlying data that are on-manifold but at its margins. This improves practical feasibility of functional outlier detection: We show that simple manifold learning methods can be used to reliably infer and visualize the geometric structure of functional data sets. We also show that standard outlier detection methods requiring tabular data inputs can be applied to functional data very successfully by simply using their vector-valued representations learned from manifold learning methods as input features. Our experiments on synthetic and real data sets demonstrate that this approach leads to outlier detection performances at least on par with existing functional data-specific methods in a large variety of settings, without the highly specialized, complex methodology and narrow domain of application these methods often entail.
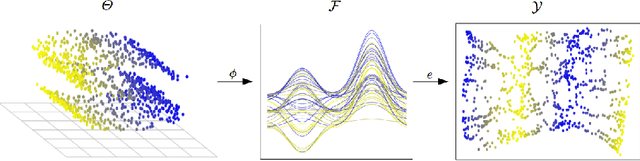
Unsupervised Functional Data Analysis via Nonlinear Dimension Reduction
Dec 22, 2020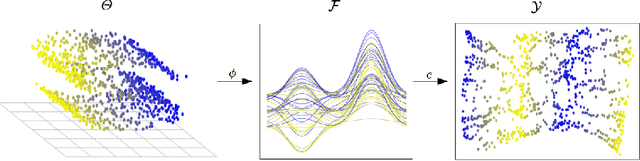
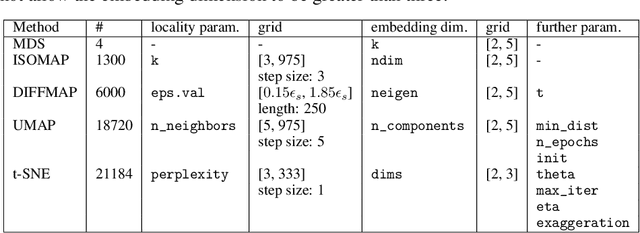
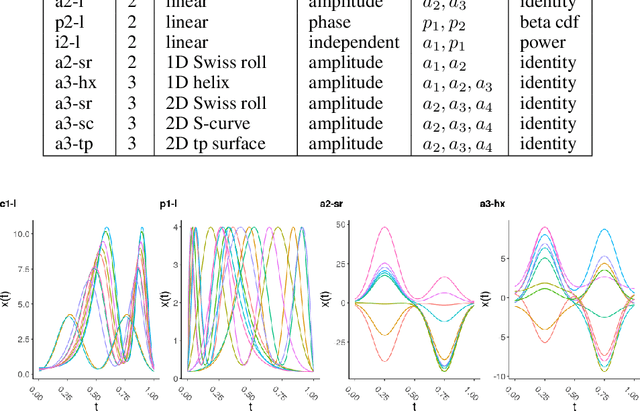
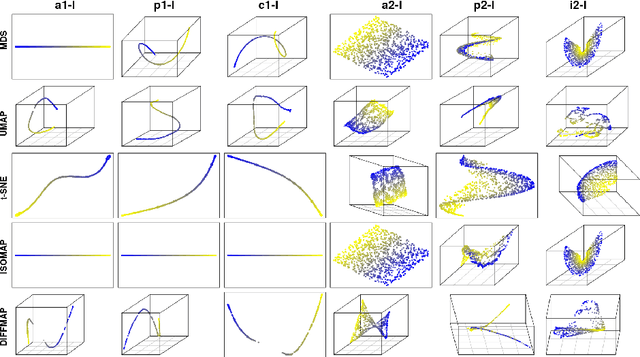
Abstract:In recent years, manifold methods have moved into focus as tools for dimension reduction. Assuming that the high-dimensional data actually lie on or close to a low-dimensional nonlinear manifold, these methods have shown convincing results in several settings. This manifold assumption is often reasonable for functional data, i.e., data representing continuously observed functions, as well. However, the performance of manifold methods recently proposed for tabular or image data has not been systematically assessed in the case of functional data yet. Moreover, it is unclear how to evaluate the quality of learned embeddings that do not yield invertible mappings, since the reconstruction error cannot be used as a performance measure for such representations. In this work, we describe and investigate the specific challenges for nonlinear dimension reduction posed by the functional data setting. The contributions of the paper are three-fold: First of all, we define a theoretical framework which allows to systematically assess specific challenges that arise in the functional data context, transfer several nonlinear dimension reduction methods for tabular and image data to functional data, and show that manifold methods can be used successfully in this setting. Secondly, we subject performance assessment and tuning strategies to a thorough and systematic evaluation based on several different functional data settings and point out some previously undescribed weaknesses and pitfalls which can jeopardize reliable judgment of embedding quality. Thirdly, we propose a nuanced approach to make trustworthy decisions for or against competing nonconforming embeddings more objectively.
Large-scale benchmark study of survival prediction methods using multi-omics data
Mar 07, 2020



Abstract:Multi-omics data, that is, datasets containing different types of high-dimensional molecular variables (often in addition to classical clinical variables), are increasingly generated for the investigation of various diseases. Nevertheless, questions remain regarding the usefulness of multi-omics data for the prediction of disease outcomes such as survival time. It is also unclear which methods are most appropriate to derive such prediction models. We aim to give some answers to these questions by means of a large-scale benchmark study using real data. Different prediction methods from machine learning and statistics were applied on 18 multi-omics cancer datasets from the database "The Cancer Genome Atlas", containing from 35 to 1,000 observations and from 60,000 to 100,000 variables. The considered outcome was the (censored) survival time. Twelve methods based on boosting, penalized regression and random forest were compared, comprising both methods that do and that do not take the group structure of the omics variables into account. The Kaplan-Meier estimate and a Cox model using only clinical variables were used as reference methods. The methods were compared using several repetitions of 5-fold cross-validation. Uno's C-index and the integrated Brier-score served as performance metrics. The results show that, although multi-omics data can improve the prediction performance, this is not generally the case. Only the method block forest slightly outperformed the Cox model on average over all datasets. Taking into account the multi-omics structure improves the predictive performance and protects variables in low-dimensional groups - especially clinical variables - from not being included in the model. All analyses are reproducible using freely available R code.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge