Tim Friede
Multi-Modal Dataset Creation for Federated~Learning with DICOM Structured Reports
Jul 12, 2024



Abstract:Purpose: Federated training is often hindered by heterogeneous datasets due to divergent data storage options, inconsistent naming schemes, varied annotation procedures, and disparities in label quality. This is particularly evident in the emerging multi-modal learning paradigms, where dataset harmonization including a uniform data representation and filtering options are of paramount importance. Methods: DICOM structured reports enable the standardized linkage of arbitrary information beyond the imaging domain and can be used within Python deep learning pipelines with highdicom. Building on this, we developed an open platform for data integration and interactive filtering capabilities that simplifies the process of assembling multi-modal datasets. Results: In this study, we extend our prior work by showing its applicability to more and divergent data types, as well as streamlining datasets for federated training within an established consortium of eight university hospitals in Germany. We prove its concurrent filtering ability by creating harmonized multi-modal datasets across all locations for predicting the outcome after minimally invasive heart valve replacement. The data includes DICOM data (i.e. computed tomography images, electrocardiography scans) as well as annotations (i.e. calcification segmentations, pointsets and pacemaker dependency), and metadata (i.e. prosthesis and diagnoses). Conclusion: Structured reports bridge the traditional gap between imaging systems and information systems. Utilizing the inherent DICOM reference system arbitrary data types can be queried concurrently to create meaningful cohorts for clinical studies. The graphical interface as well as example structured report templates will be made publicly available.
Federated Foundation Model for Cardiac CT Imaging
Jul 10, 2024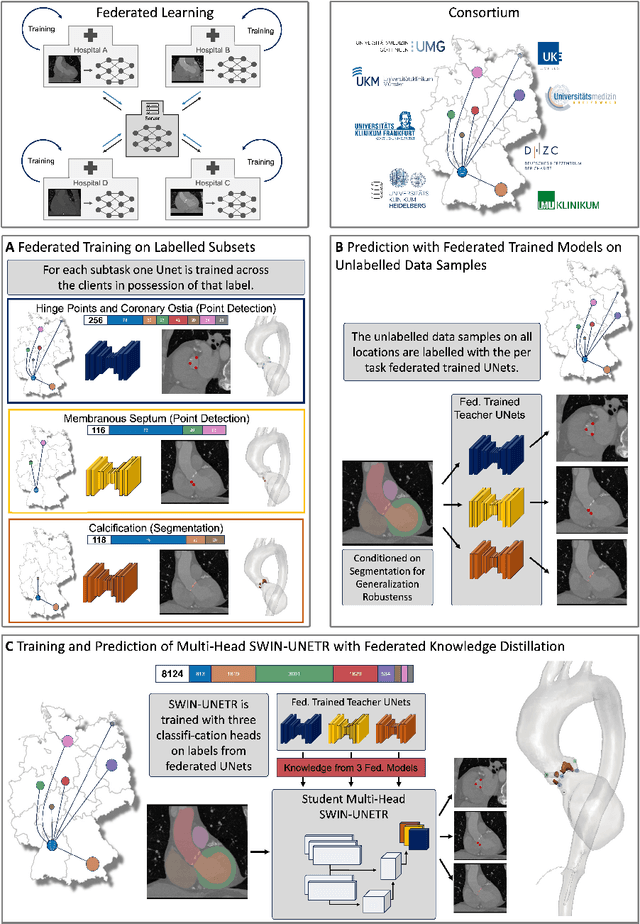
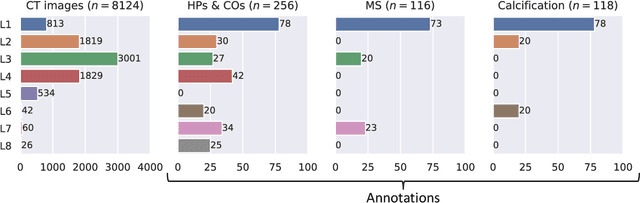
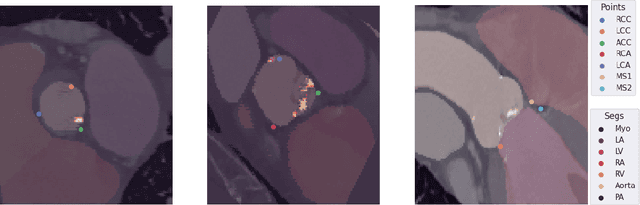

Abstract:Federated learning (FL) is a renowned technique for utilizing decentralized data while preserving privacy. However, real-world applications often involve inherent challenges such as partially labeled datasets, where not all clients possess expert annotations of all labels of interest, leaving large portions of unlabeled data unused. In this study, we conduct the largest federated cardiac CT imaging analysis to date, focusing on partially labeled datasets ($n=8,124$) of Transcatheter Aortic Valve Implantation (TAVI) patients over eight hospital clients. Transformer architectures, which are the major building blocks of current foundation models, have shown superior performance when trained on larger cohorts than traditional CNNs. However, when trained on small task-specific labeled sample sizes, it is currently not feasible to exploit their underlying attention mechanism for improved performance. Therefore, we developed a two-stage semi-supervised learning strategy that distills knowledge from several task-specific CNNs (landmark detection and segmentation of calcification) into a single transformer model by utilizing large amounts of unlabeled data typically residing unused in hospitals to mitigate these issues. This method not only improves the predictive accuracy and generalizability of transformer-based architectures but also facilitates the simultaneous learning of all partial labels within a single transformer model across the federation. Additionally, we show that our transformer-based model extracts more meaningful features for further downstream tasks than the UNet-based one by only training the last layer to also solve segmentation of coronary arteries. We make the code and weights of the final model openly available, which can serve as a foundation model for further research in cardiac CT imaging.
Model-based recursive partitioning for discrete event times
Sep 14, 2022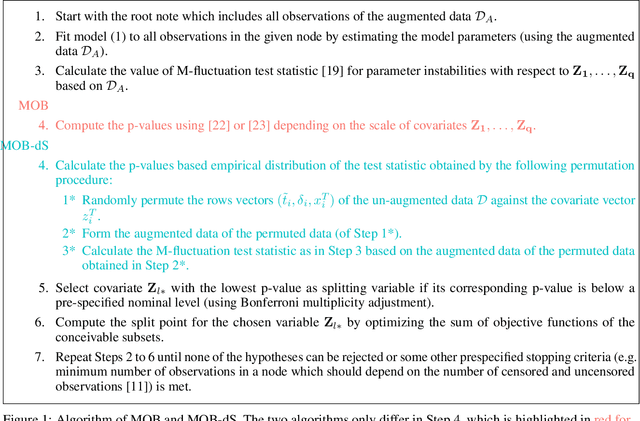

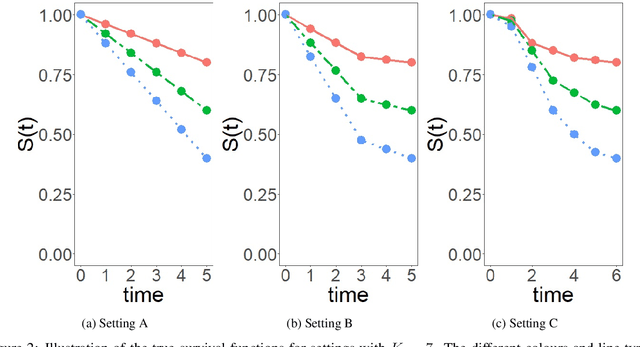
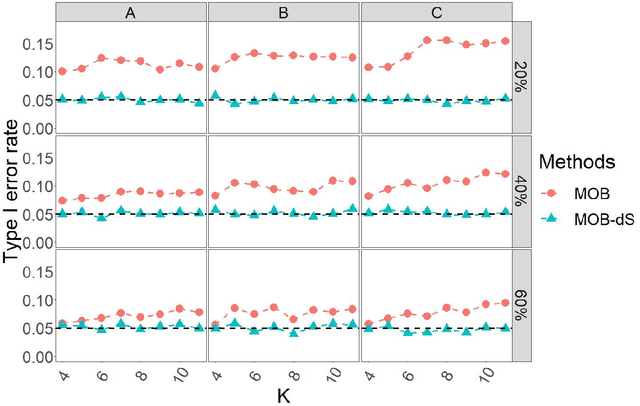
Abstract:Model-based recursive partitioning (MOB) is a semi-parametric statistical approach allowing the identification of subgroups that can be combined with a broad range of outcome measures including continuous time-to-event outcomes. When time is measured on a discrete scale, methods and models need to account for this discreetness as otherwise subgroups might be spurious and effects biased. The test underlying the splitting criterion of MOB, the M-fluctuation test, assumes independent observations. However, for fitting discrete time-to-event models the data matrix has to be modified resulting in an augmented data matrix violating the independence assumption. We propose MOB for discrete Survival data (MOB-dS) which controls the type I error rate of the test used for data splitting and therefore the rate of identifying subgroups although none is present. MOB-ds uses a permutation approach accounting for dependencies in the augmented time-to-event data to obtain the distribution under the null hypothesis of no subgroups being present. Through simulations we investigate the type I error rate of the new MOB-dS and the standard MOB for different patterns of survival curves and event rates. We find that the type I error rates of the test is well controlled for MOB-dS, but observe some considerable inflations of the error rate for MOB. To illustrate the proposed methods, MOB-dS is applied to data on unemployment duration.
On the role of benchmarking data sets and simulations in method comparison studies
Aug 02, 2022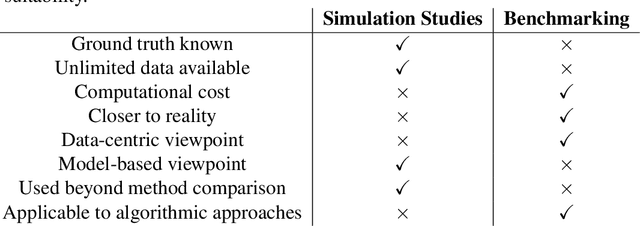
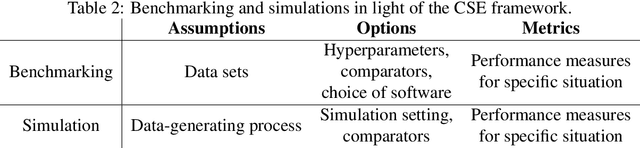
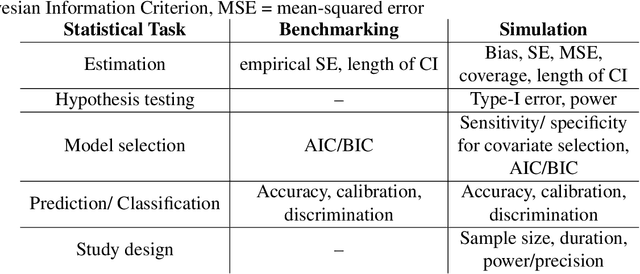
Abstract:Method comparisons are essential to provide recommendations and guidance for applied researchers, who often have to choose from a plethora of available approaches. While many comparisons exist in the literature, these are often not neutral but favour a novel method. Apart from the choice of design and a proper reporting of the findings, there are different approaches concerning the underlying data for such method comparison studies. Most manuscripts on statistical methodology rely on simulation studies and provide a single real-world data set as an example to motivate and illustrate the methodology investigated. In the context of supervised learning, in contrast, methods are often evaluated using so-called benchmarking data sets, i.e. real-world data that serve as gold standard in the community. Simulation studies, on the other hand, are much less common in this context. The aim of this paper is to investigate differences and similarities between these approaches, to discuss their advantages and disadvantages and ultimately to develop new approaches to the evaluation of methods picking the best of both worlds. To this aim, we borrow ideas from different contexts such as mixed methods research and Clinical Scenario Evaluation.
Is there a role for statistics in artificial intelligence?
Sep 13, 2020

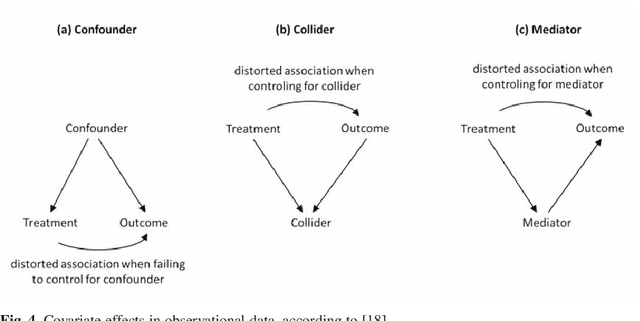
Abstract:The research on and application of artificial intelligence (AI) has triggered a comprehensive scientific, economic, social and political discussion. Here we argue that statistics, as an interdisciplinary scientific field, plays a substantial role both for the theoretical and practical understanding of AI and for its future development. Statistics might even be considered a core element of AI. With its specialist knowledge of data evaluation, starting with the precise formulation of the research question and passing through a study design stage on to analysis and interpretation of the results, statistics is a natural partner for other disciplines in teaching, research and practice. This paper aims at contributing to the current discussion by highlighting the relevance of statistical methodology in the context of AI development. In particular, we discuss contributions of statistics to the field of artificial intelligence concerning methodological development, planning and design of studies, assessment of data quality and data collection, differentiation of causality and associations and assessment of uncertainty in results. Moreover, the paper also deals with the equally necessary and meaningful extension of curricula in schools and universities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge