Martin Ester
Compositional Flows for 3D Molecule and Synthesis Pathway Co-design
Apr 10, 2025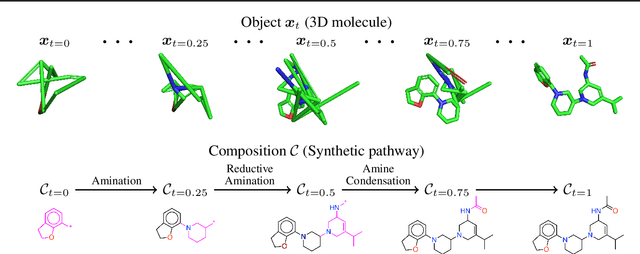
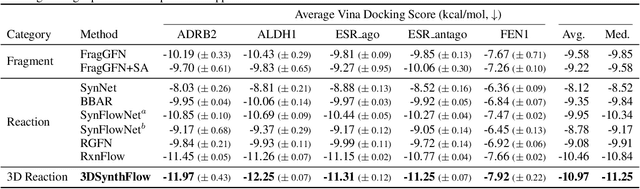
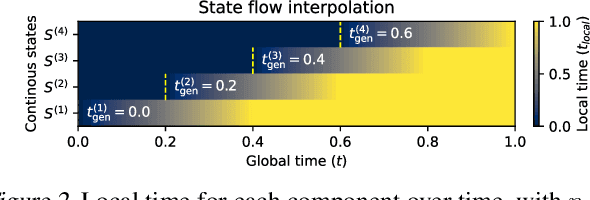
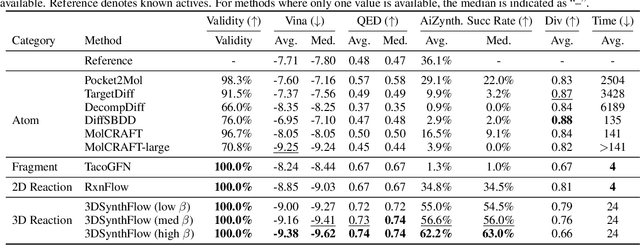
Abstract:Many generative applications, such as synthesis-based 3D molecular design, involve constructing compositional objects with continuous features. Here, we introduce Compositional Generative Flows (CGFlow), a novel framework that extends flow matching to generate objects in compositional steps while modeling continuous states. Our key insight is that modeling compositional state transitions can be formulated as a straightforward extension of the flow matching interpolation process. We further build upon the theoretical foundations of generative flow networks (GFlowNets), enabling reward-guided sampling of compositional structures. We apply CGFlow to synthesizable drug design by jointly designing the molecule's synthetic pathway with its 3D binding pose. Our approach achieves state-of-the-art binding affinity on all 15 targets from the LIT-PCBA benchmark, and 5.8$\times$ improvement in sampling efficiency compared to 2D synthesis-based baseline. To our best knowledge, our method is also the first to achieve state of-art-performance in both Vina Dock (-9.38) and AiZynth success rate (62.2\%) on the CrossDocked benchmark.
Improving OOD Generalization of Pre-trained Encoders via Aligned Embedding-Space Ensembles
Nov 20, 2024



Abstract:The quality of self-supervised pre-trained embeddings on out-of-distribution (OOD) data is poor without fine-tuning. A straightforward and simple approach to improving the generalization of pre-trained representation to OOD data is the use of deep ensembles. However, obtaining an effective ensemble in the embedding space with only unlabeled data remains an unsolved problem. We first perform a theoretical analysis that reveals the relationship between individual hyperspherical embedding spaces in an ensemble. We then design a principled method to align these embedding spaces in an unsupervised manner. Experimental results on the MNIST dataset show that our embedding-space ensemble method improves pre-trained embedding quality on in-distribution and OOD data compared to single encoders.
Causal Order Discovery based on Monotonic SCMs
Oct 24, 2024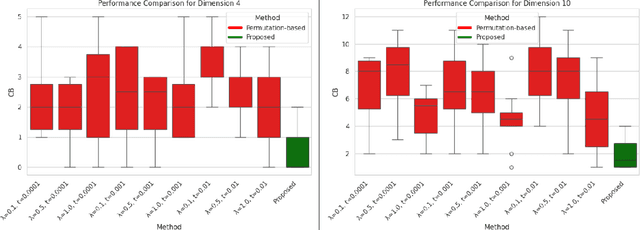
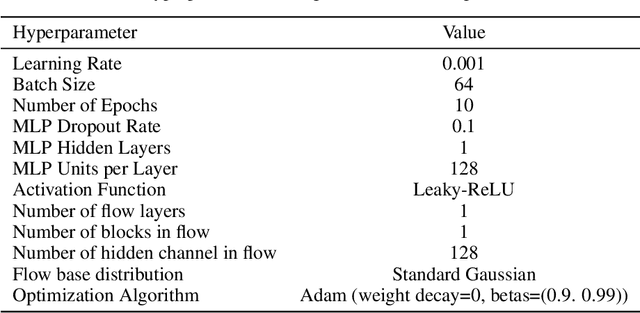
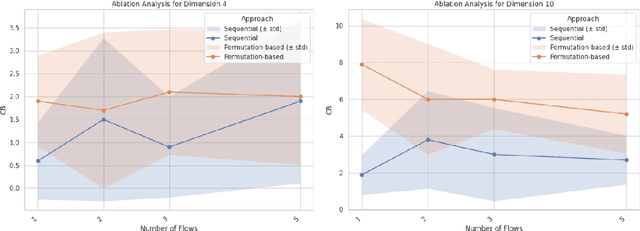
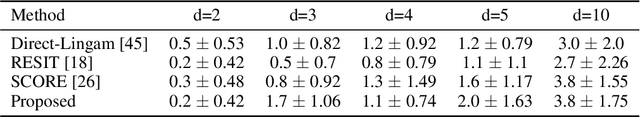
Abstract:In this paper, we consider the problem of causal order discovery within the framework of monotonic Structural Causal Models (SCMs), which have gained attention for their potential to enable causal inference and causal discovery from observational data. While existing approaches either assume prior knowledge about the causal order or use complex optimization techniques to impose sparsity in the Jacobian of Triangular Monotonic Increasing maps, our work introduces a novel sequential procedure that directly identifies the causal order by iteratively detecting the root variable. This method eliminates the need for sparsity assumptions and the associated optimization challenges, enabling the identification of a unique SCM without the need for multiple independence tests to break the Markov equivalence class. We demonstrate the effectiveness of our approach in sequentially finding the root variable, comparing it to methods that maximize Jacobian sparsity.
Generative Flows on Synthetic Pathway for Drug Design
Oct 06, 2024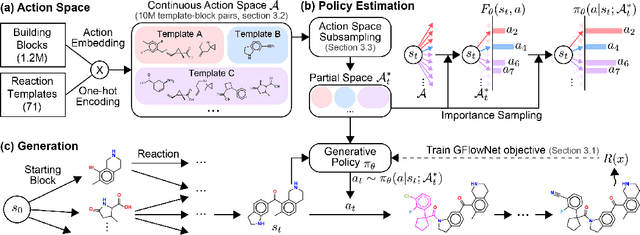
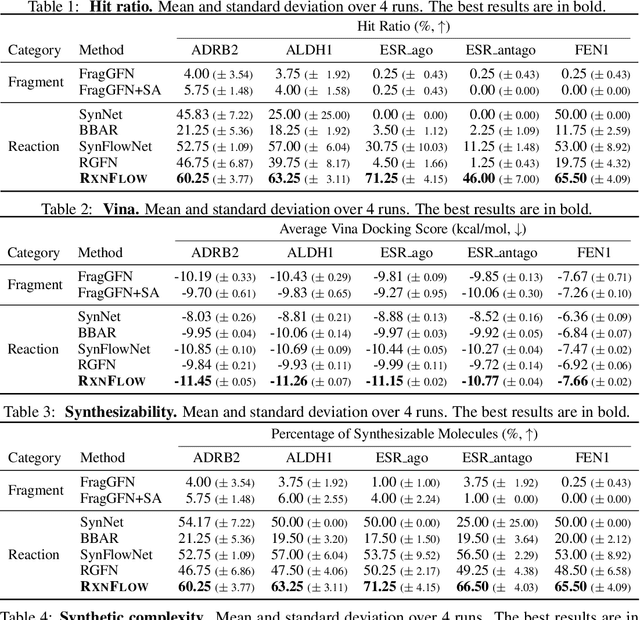
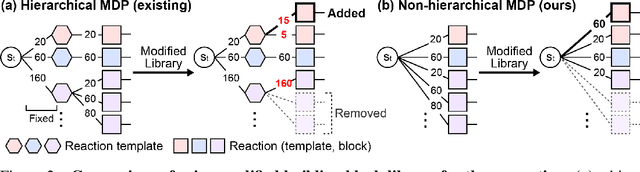
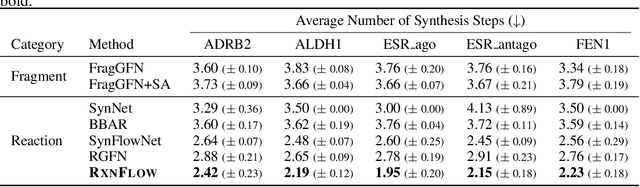
Abstract:Generative models in drug discovery have recently gained attention as efficient alternatives to brute-force virtual screening. However, most existing models do not account for synthesizability, limiting their practical use in real-world scenarios. In this paper, we propose RxnFlow, which sequentially assembles molecules using predefined molecular building blocks and chemical reaction templates to constrain the synthetic chemical pathway. We then train on this sequential generating process with the objective of generative flow networks (GFlowNets) to generate both highly rewarded and diverse molecules. To mitigate the large action space of synthetic pathways in GFlowNets, we implement a novel action space subsampling method. This enables RxnFlow to learn generative flows over extensive action spaces comprising combinations of 1.2 million building blocks and 71 reaction templates without significant computational overhead. Additionally, RxnFlow can employ modified or expanded action spaces for generation without retraining, allowing for the introduction of additional objectives or the incorporation of newly discovered building blocks. We experimentally demonstrate that RxnFlow outperforms existing reaction-based and fragment-based models in pocket-specific optimization across various target pockets. Furthermore, RxnFlow achieves state-of-the-art performance on CrossDocked2020 for pocket-conditional generation, with an average Vina score of -8.85kcal/mol and 34.8% synthesizability.
UnPaSt: unsupervised patient stratification by differentially expressed biclusters in omics data
Jul 31, 2024Abstract:Most complex diseases, including cancer and non-malignant diseases like asthma, have distinct molecular subtypes that require distinct clinical approaches. However, existing computational patient stratification methods have been benchmarked almost exclusively on cancer omics data and only perform well when mutually exclusive subtypes can be characterized by many biomarkers. Here, we contribute with a massive evaluation attempt, quantitatively exploring the power of 22 unsupervised patient stratification methods using both, simulated and real transcriptome data. From this experience, we developed UnPaSt (https://apps.cosy.bio/unpast/) optimizing unsupervised patient stratification, working even with only a limited number of subtype-predictive biomarkers. We evaluated all 23 methods on real-world breast cancer and asthma transcriptomics data. Although many methods reliably detected major breast cancer subtypes, only few identified Th2-high asthma, and UnPaSt significantly outperformed its closest competitors in both test datasets. Essentially, we showed that UnPaSt can detect many biologically insightful and reproducible patterns in omic datasets.
Geometric-informed GFlowNets for Structure-Based Drug Design
Jun 16, 2024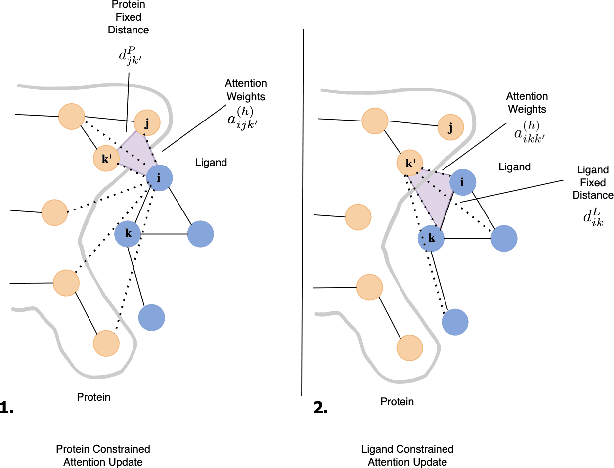


Abstract:The rise of cost involved with drug discovery and current speed of which they are discover, underscore the need for more efficient structure-based drug design (SBDD) methods. We employ Generative Flow Networks (GFlowNets), to effectively explore the vast combinatorial space of drug-like molecules, which traditional virtual screening methods fail to cover. We introduce a novel modification to the GFlowNet framework by incorporating trigonometrically consistent embeddings, previously utilized in tasks involving protein conformation and protein-ligand interactions, to enhance the model's ability to generate molecules tailored to specific protein pockets. We have modified the existing protein conditioning used by GFlowNets, blending geometric information from both protein and ligand embeddings to achieve more geometrically consistent embeddings. Experiments conducted using CrossDocked2020 demonstrated an improvement in the binding affinity between generated molecules and protein pockets for both single and multi-objective tasks, compared to previous work. Additionally, we propose future work aimed at further increasing the geometric information captured in protein-ligand interactions.
IceFormer: Accelerated Inference with Long-Sequence Transformers on CPUs
May 05, 2024



Abstract:One limitation of existing Transformer-based models is that they cannot handle very long sequences as input since their self-attention operations exhibit quadratic time and space complexity. This problem becomes especially acute when Transformers are deployed on hardware platforms equipped only with CPUs. To address this issue, we propose a novel method for accelerating self-attention at inference time that works with pretrained Transformer models out-of-the-box without requiring retraining. We experiment using our method to accelerate various long-sequence Transformers, including a leading LLaMA 2-based LLM, on various benchmarks and demonstrate a greater speedup of 2.73x - 7.63x while retaining 98.6% - 99.6% of the accuracy of the original pretrained models. The code is available on our project website at https://yuzhenmao.github.io/IceFormer/.
Adversarially Balanced Representation for Continuous Treatment Effect Estimation
Dec 17, 2023Abstract:Individual treatment effect (ITE) estimation requires adjusting for the covariate shift between populations with different treatments, and deep representation learning has shown great promise in learning a balanced representation of covariates. However the existing methods mostly consider the scenario of binary treatments. In this paper, we consider the more practical and challenging scenario in which the treatment is a continuous variable (e.g. dosage of a medication), and we address the two main challenges of this setup. We propose the adversarial counterfactual regression network (ACFR) that adversarially minimizes the representation imbalance in terms of KL divergence, and also maintains the impact of the treatment value on the outcome prediction by leveraging an attention mechanism. Theoretically we demonstrate that ACFR objective function is grounded in an upper bound on counterfactual outcome prediction error. Our experimental evaluation on semi-synthetic datasets demonstrates the empirical superiority of ACFR over a range of state-of-the-art methods.
TacoGFN: Target Conditioned GFlowNet for Structure-Based Drug Design
Oct 05, 2023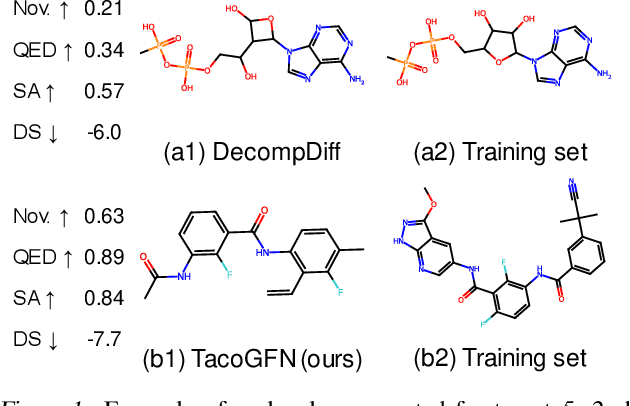



Abstract:We seek to automate the generation of drug-like compounds conditioned to specific protein pocket targets. Most current methods approximate the protein-molecule distribution of a finite dataset and, therefore struggle to generate molecules with significant binding improvement over the training dataset. We instead frame the pocket-conditioned molecular generation task as an RL problem and develop TacoGFN, a target conditional Generative Flow Network model. Our method is explicitly encouraged to generate molecules with desired properties as opposed to fitting on a pre-existing data distribution. To this end, we develop transformer-based docking score prediction to speed up docking score computation and propose TacoGFN to explore molecule space efficiently. Furthermore, we incorporate several rounds of active learning where generated samples are queried using a docking oracle to improve the docking score prediction. This approach allows us to accurately explore as much of the molecule landscape as we can afford computationally. Empirically, molecules generated using TacoGFN and its variants significantly outperform all baseline methods across every property (Docking score, QED, SA, Lipinski), while being orders of magnitude faster.
Semi-Supervised Junction Tree Variational Autoencoder for Molecular Property Prediction
Sep 01, 2022

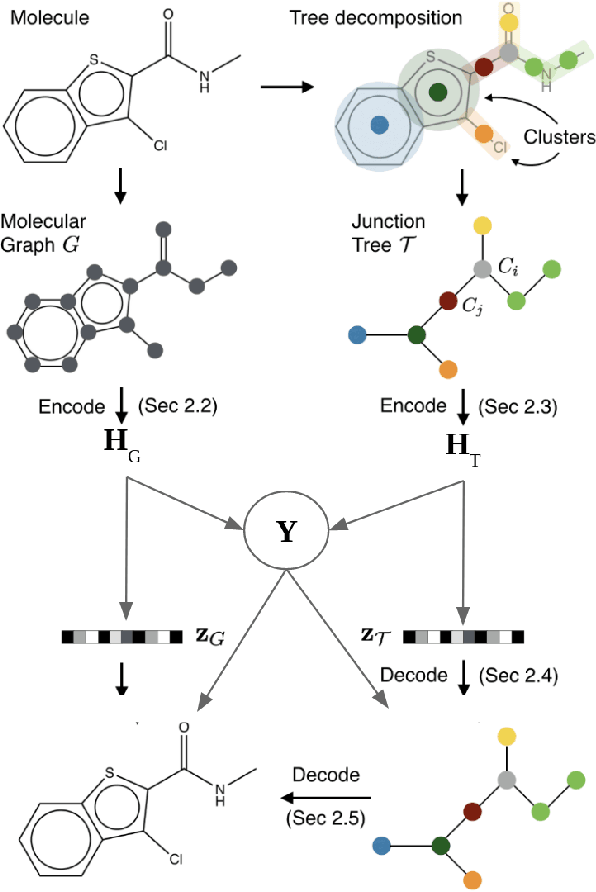
Abstract:Recent advances in machine learning have enabled accurate prediction of chemical properties. However, supervised machine learning methods in this domain often suffer from the label scarcity problem, due to the expensive nature of labeling chemical property experimentally. This research modifies state-of-the-art molecule generation method - Junction Tree Variational Autoencoder (JT-VAE) to facilitate semi-supervised learning on chemical property prediction. Furthermore, we force some latent variables to take on consistent and interpretable purposes such as representing toxicity via this partial supervision. We leverage JT-VAE architecture to learn an interpretable representation optimal for tasks ranging from molecule property prediction to conditional molecule generation, using a partially labelled dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge