Semi-Supervised Junction Tree Variational Autoencoder for Molecular Property Prediction
Paper and Code
Sep 01, 2022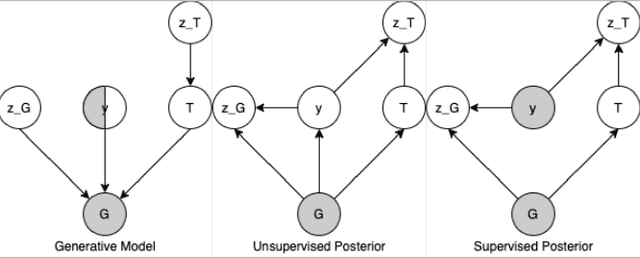

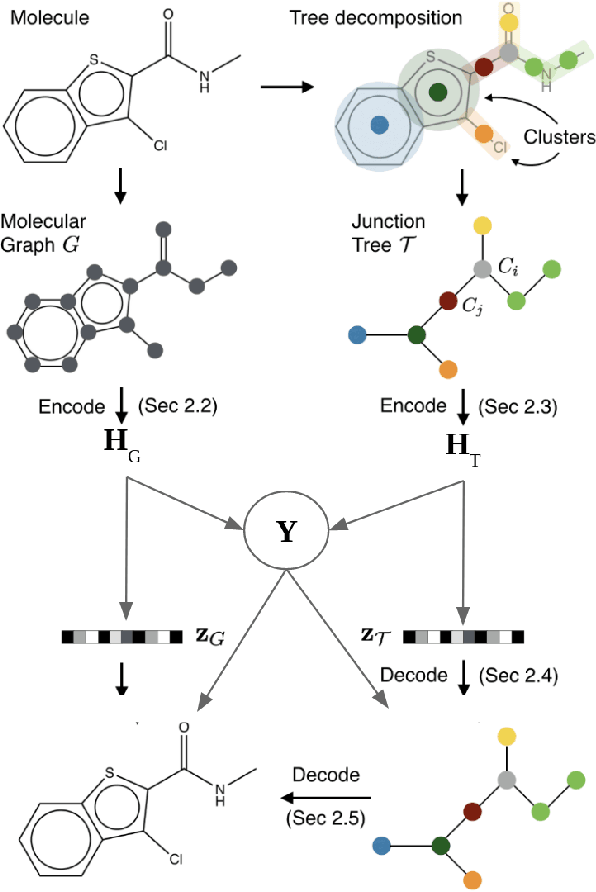
Recent advances in machine learning have enabled accurate prediction of chemical properties. However, supervised machine learning methods in this domain often suffer from the label scarcity problem, due to the expensive nature of labeling chemical property experimentally. This research modifies state-of-the-art molecule generation method - Junction Tree Variational Autoencoder (JT-VAE) to facilitate semi-supervised learning on chemical property prediction. Furthermore, we force some latent variables to take on consistent and interpretable purposes such as representing toxicity via this partial supervision. We leverage JT-VAE architecture to learn an interpretable representation optimal for tasks ranging from molecule property prediction to conditional molecule generation, using a partially labelled dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge