Atia Hamidizadeh
Mem2Ego: Empowering Vision-Language Models with Global-to-Ego Memory for Long-Horizon Embodied Navigation
Feb 20, 2025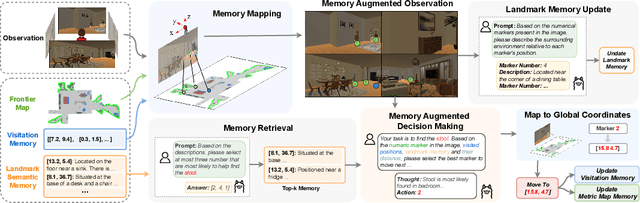
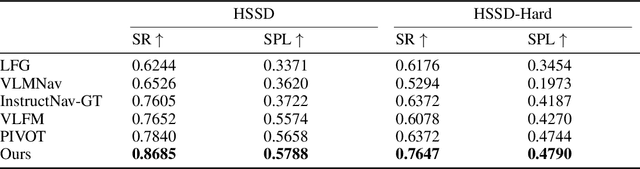
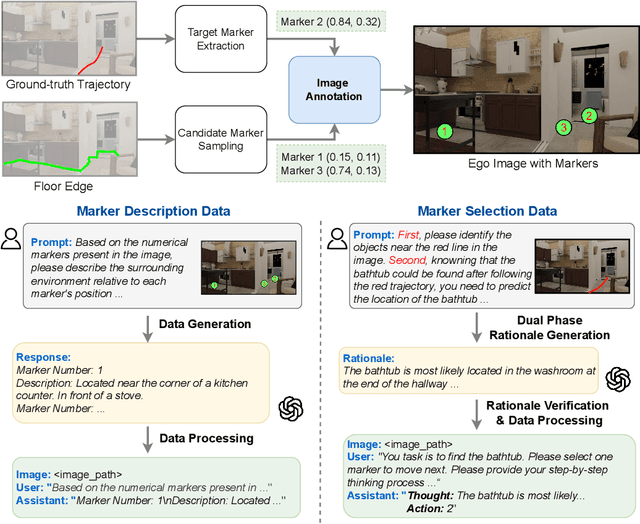
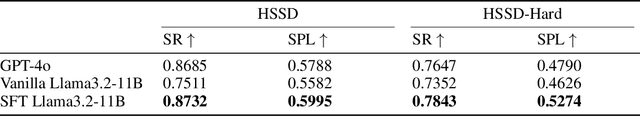
Abstract:Recent advancements in Large Language Models (LLMs) and Vision-Language Models (VLMs) have made them powerful tools in embodied navigation, enabling agents to leverage commonsense and spatial reasoning for efficient exploration in unfamiliar environments. Existing LLM-based approaches convert global memory, such as semantic or topological maps, into language descriptions to guide navigation. While this improves efficiency and reduces redundant exploration, the loss of geometric information in language-based representations hinders spatial reasoning, especially in intricate environments. To address this, VLM-based approaches directly process ego-centric visual inputs to select optimal directions for exploration. However, relying solely on a first-person perspective makes navigation a partially observed decision-making problem, leading to suboptimal decisions in complex environments. In this paper, we present a novel vision-language model (VLM)-based navigation framework that addresses these challenges by adaptively retrieving task-relevant cues from a global memory module and integrating them with the agent's egocentric observations. By dynamically aligning global contextual information with local perception, our approach enhances spatial reasoning and decision-making in long-horizon tasks. Experimental results demonstrate that the proposed method surpasses previous state-of-the-art approaches in object navigation tasks, providing a more effective and scalable solution for embodied navigation.
E2ESlack: An End-to-End Graph-Based Framework for Pre-Routing Slack Prediction
Jan 14, 2025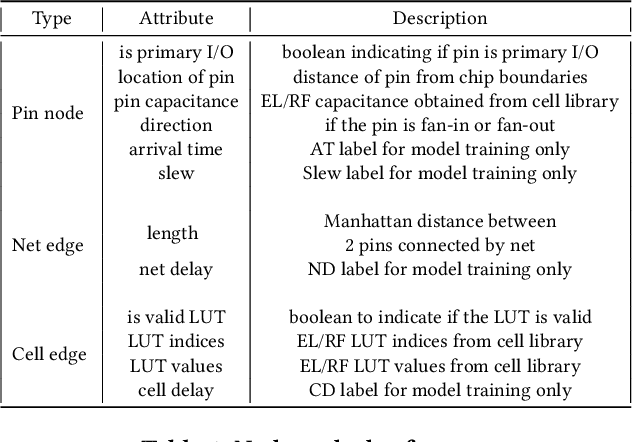
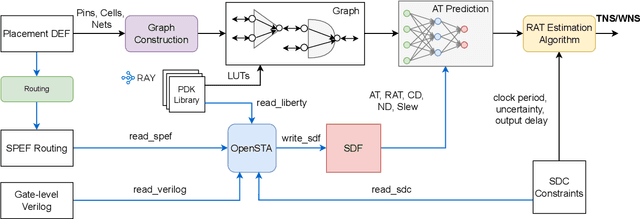
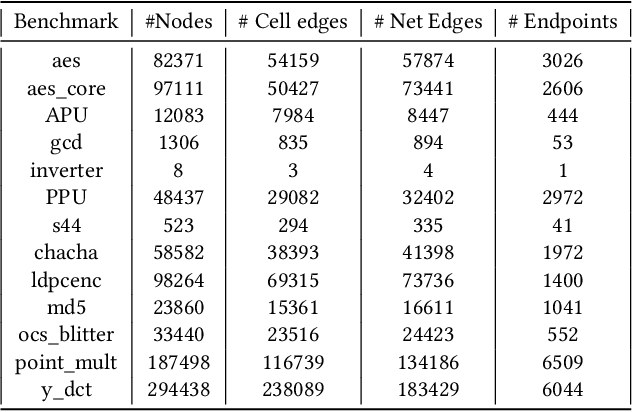
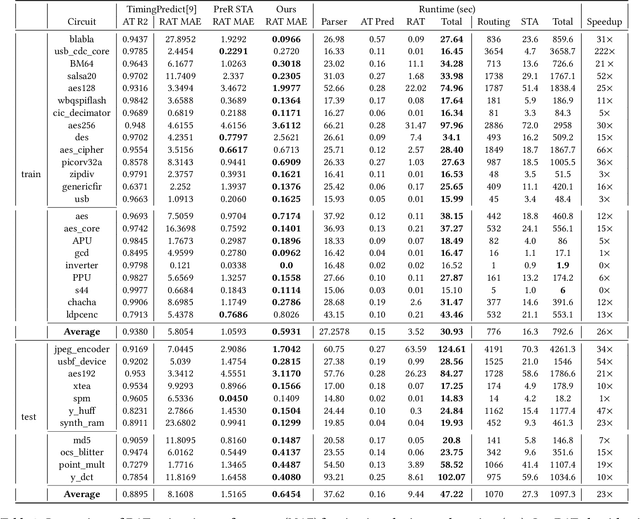
Abstract:Pre-routing slack prediction remains a critical area of research in Electronic Design Automation (EDA). Despite numerous machine learning-based approaches targeting this task, there is still a lack of a truly end-to-end framework that engineers can use to obtain TNS/WNS metrics from raw circuit data at the placement stage. Existing works have demonstrated effectiveness in Arrival Time (AT) prediction but lack a mechanism for Required Arrival Time (RAT) prediction, which is essential for slack prediction and obtaining TNS/WNS metrics. In this work, we propose E2ESlack, an end-to-end graph-based framework for pre-routing slack prediction. The framework includes a TimingParser that supports DEF, SDF and LIB files for feature extraction and graph construction, an arrival time prediction model and a fast RAT estimation module. To the best of our knowledge, this is the first work capable of predicting path-level slacks at the pre-routing stage. We perform extensive experiments and demonstrate that our proposed RAT estimation method outperforms the SOTA ML-based prediction method and also pre-routing STA tool. Additionally, the proposed E2ESlack framework achieves TNS/WNS values comparable to post-routing STA results while saving up to 23x runtime.
Semi-Supervised Junction Tree Variational Autoencoder for Molecular Property Prediction
Sep 01, 2022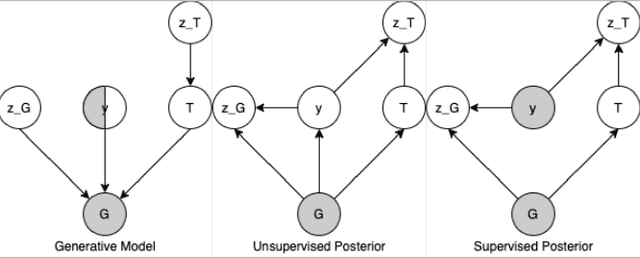

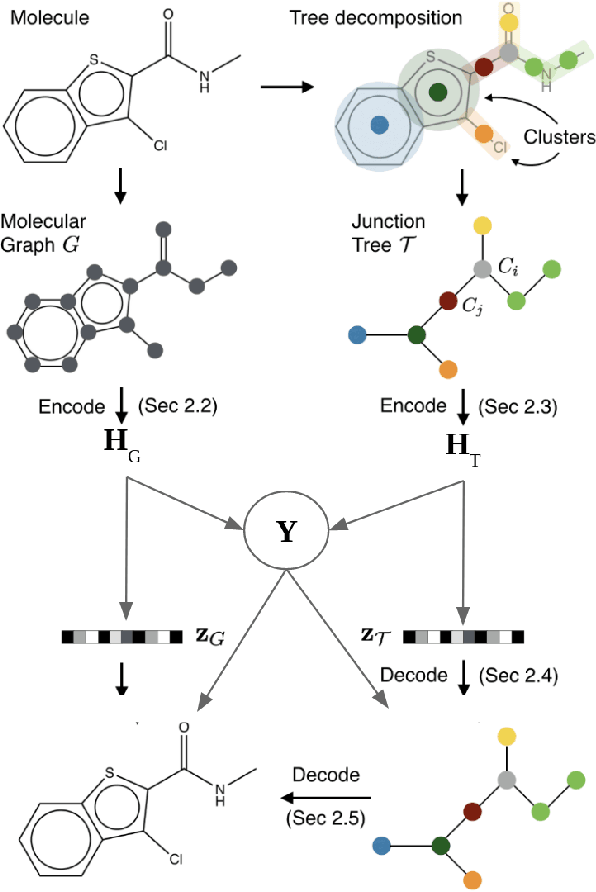
Abstract:Recent advances in machine learning have enabled accurate prediction of chemical properties. However, supervised machine learning methods in this domain often suffer from the label scarcity problem, due to the expensive nature of labeling chemical property experimentally. This research modifies state-of-the-art molecule generation method - Junction Tree Variational Autoencoder (JT-VAE) to facilitate semi-supervised learning on chemical property prediction. Furthermore, we force some latent variables to take on consistent and interpretable purposes such as representing toxicity via this partial supervision. We leverage JT-VAE architecture to learn an interpretable representation optimal for tasks ranging from molecule property prediction to conditional molecule generation, using a partially labelled dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge